Draft reconstruction
generation
Firstly, you need to have a close related model
organism whose metabolic reconstruction has been built already.
Then you need to prepare a blank reconstruction
containing gene information of your interest strain. This file can be generated
by GBKParser (http://sb.nhri.org.tw/GEMSiRV/en/GBKParser).
However, you need to add the corresponding orthologous genes to the column of Ref-BLAST.
For example, we want to draft a reconstruction of Salmonella enteric subsp. Enteric
serovar Typhimurium str.
LT2 (SLT2) by mapping to the reconstruction model iAF1260 of Escherichia coli
str. K-12 substr. MG1655 (ECO).
Therefore, we download the gbk
files of these two strains from RefSeq (http://www.ncbi.nlm.nih.gov/RefSeq).
With available NC_003197.gbk and NC_000913.gbk files for SLT2 and ECO
respectively, we then use GBKParser to parse basic
gene information and amino acid sequences. In addition, we download the
metabolic model iAF1260 from BiGG (http://bigg.ucsd.edu/)
and modify it with TextReplacer (http://sb.nhri.org.tw/GEMSiRV/en/TextReplacer).
The ready-to-use model can be found and downloaded from http://sb.nhri.org.tw/GEMSiRV/en/Metabolic_Models.
The amino acid sequence files for SLT2 and ECO can be
used to generate the reciprocal orthologous-gene
pairs by BLASTP or other available software. For example, MrBac
(http://sb.nhri.org.tw/MrBac) can be
used to generate the needed file. However, the detailed procedure is not described
here.
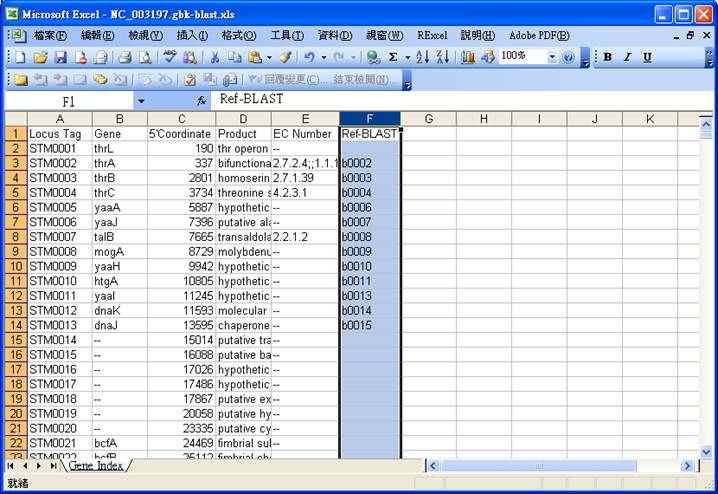
The basic gene information parsed from the gbk file is outputted to a spreadsheet file, e.g.
NC_003197.gbk.xls, which can be imported into GEMSiRV
directly. Right click on Model Databases
to Import spreadsheet (.xls).
Original
spreadsheet file:
Required fields
![]()
![]()
![]()
![]()
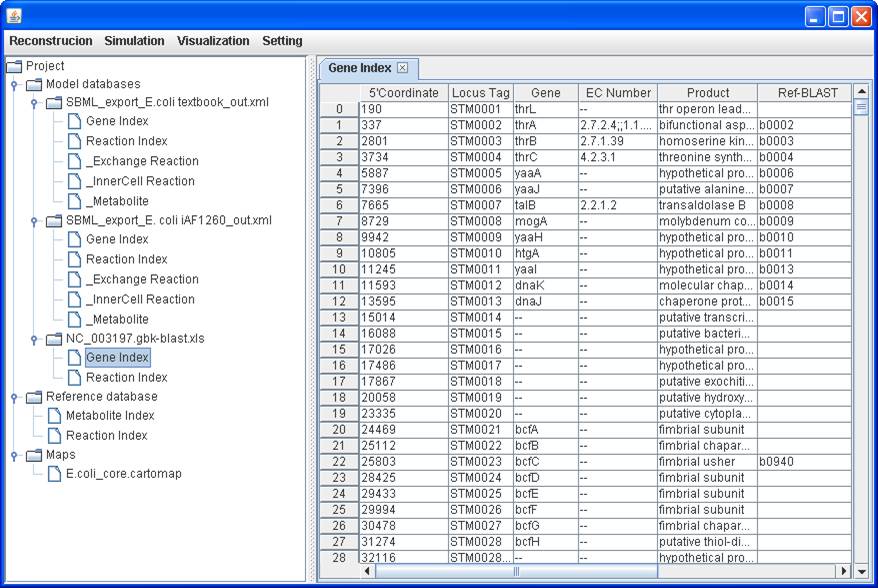
Gene Index table
of the imported blank reconstruction (NC_003197.gbk-blast.xls):
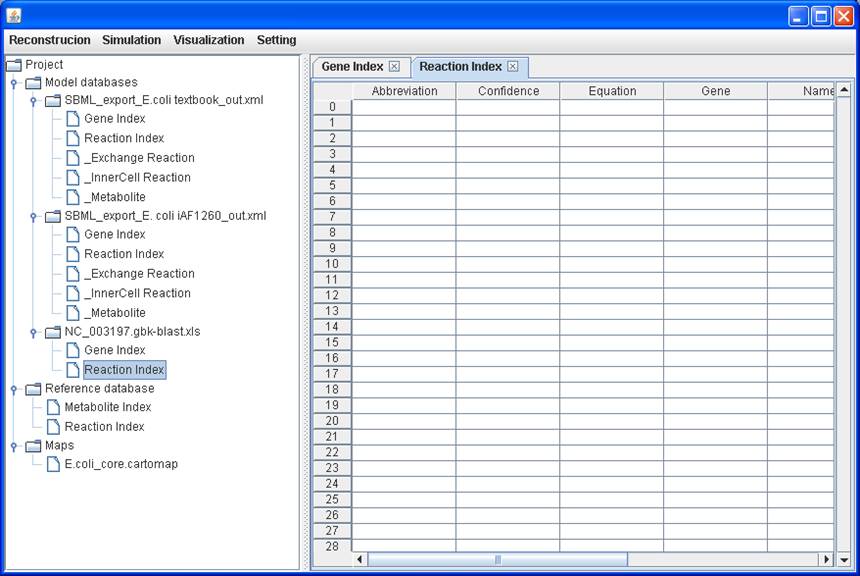
Empty Reaction
Index table of the imported blank reconstruction:
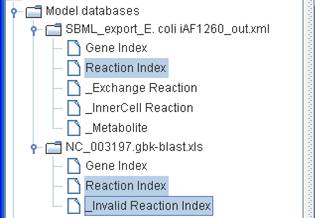
Right click
on the blank reconstruction to Draft a reconstruction by choosing SBML_export_E.coli iAF1260_out.xml
as the reference model.
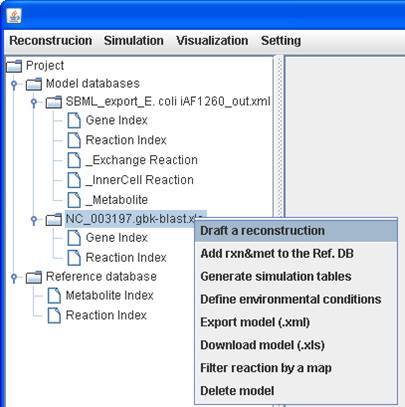
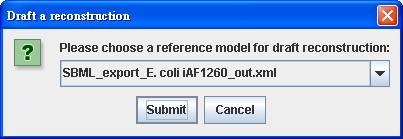
The reactions
in the reference reconstruction are classified into two indices (Reaction Index
and _Invalid Reaction Index) for the draft reconstruction: one list containing
reactions whose associated orthologous genes are present
in the blank reconstruction and conform to Boolean statements as described in
the reference reconstruction, the other containing those reactions with unknown
gene-reaction associations or reactions whose orthologous
genes are absent and let to disagree Boolean statements.