Model refinement
Based on the
draft reconstruction generated from Model SEED (http://seed-viewer.theseed.org/seedviewer.cgi?page=ModelView)
or GEMSiRV, users can curate and refine the reconstruction in GEMSiRV. However,
the lack of gene information in imported models may hinder the progress. We, therefore,
provide a function to load and update the gene information in GEMSiRV. You can
right click on the Gene Index of a model to Load and update gene’s info.,
and upload the spreadsheet file generated by GBKPaser (http://sb.nhri.org.tw/GEMSiRV/en/GBKParser),
e.g. NC_000913.gbk.xls for ECO.
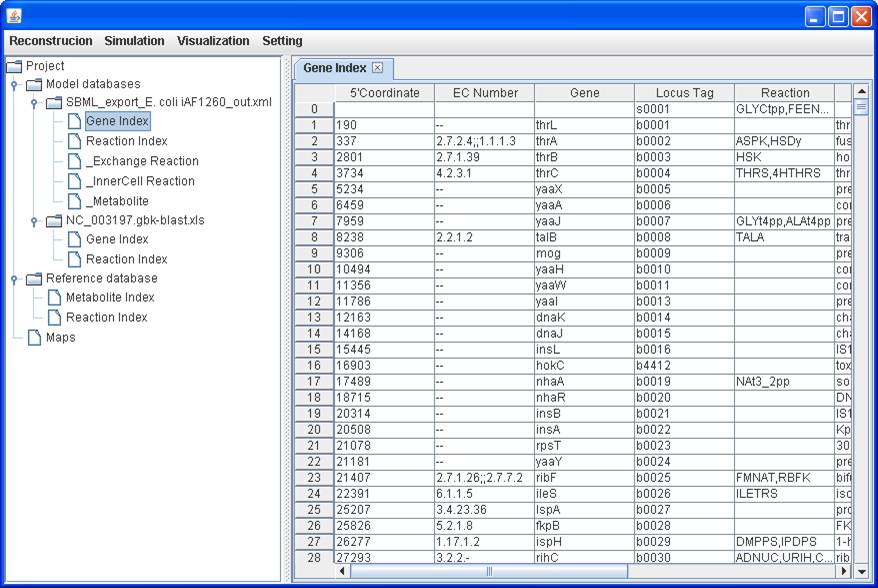
Gene information can be loaded and updated accordingly:
With the aids
of simulation and visualization, users can readily identify dead-end
metabolites and blocked reactions in the models. Prior to perform simulation,
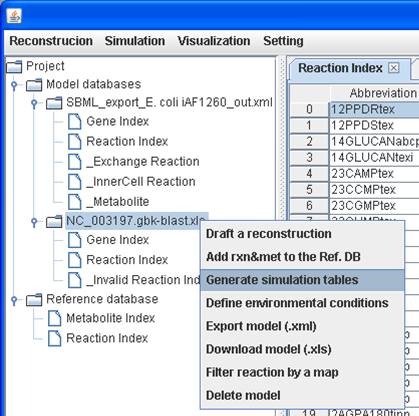
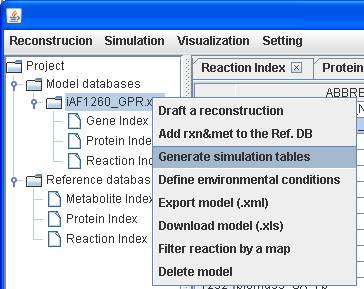
users need to convert the reconstruction into a mathematical model. Therefore,
you can right click on a model to Generate simulation tables to generate
a model containing a stoichiometric matrix as well as default systems
boundaries.
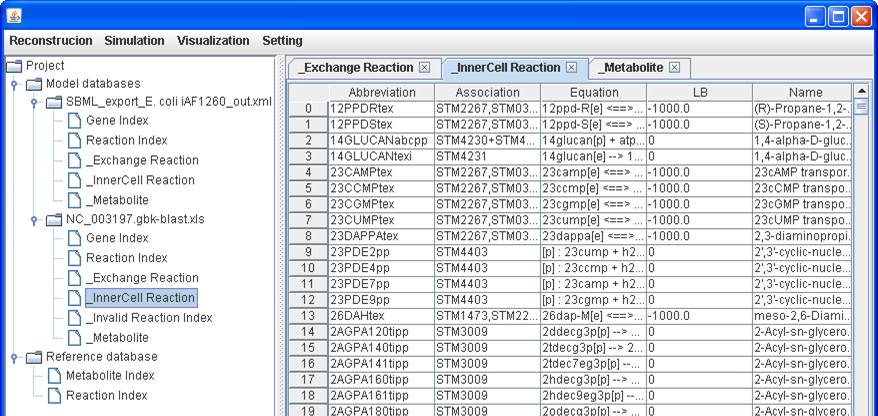
After
clicking on Generate simulation tables, three tables including InnerCell
Reaction, Exchange Reaction and Metabolite are generated. The prefix “_” used in
these three tables for easily distinguishing from the tables required for
reconstruction, e.g. Gene Index, Protein Index (optional) and Reaction Index.

_InnerCell
Reaction:
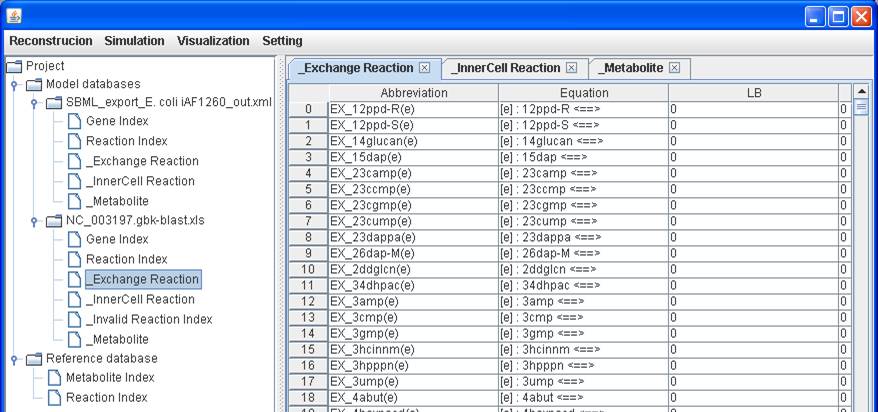
_Exchange
Reaction:
Please note
that the _Exchange Reaction table will be generated only when you have
exchanging metabolites (i.e. extracellular metabolites) in the reaction
equations.
_Metabolite:
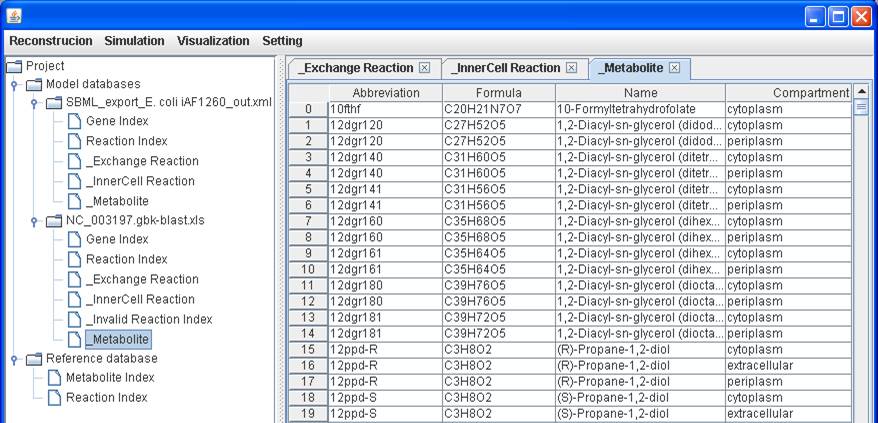
Because growth
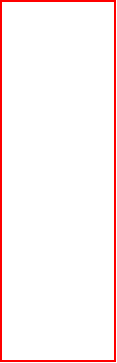
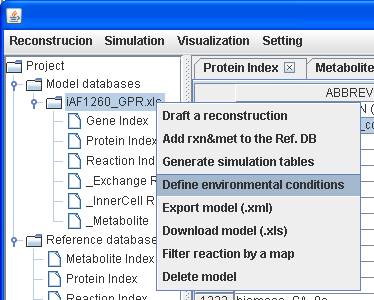
media for modeled organisms may be similar, an environmental condition can be
easily set to a model by right clicking on the model to Define environmental
conditions.
Here we use
the in silico (computational) minimal
media for the model iAF1260 as an
example (the text file can be downloaded in http://sb.nhri.org.tw/GEMSiRV/en/Manual).
In order to set the system boundaries to the default values, we right click on
the model to Generate simulation tables.
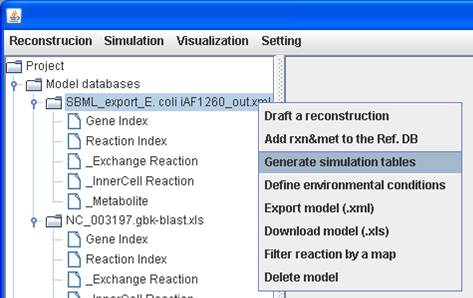
The new
simulation tables are generated and replace the previous tables. We set a
growth medium for modeling the model. We prepare a text file containing the user-defined
boundaries and objective, and then right click on the model to Define
environmental conditions.
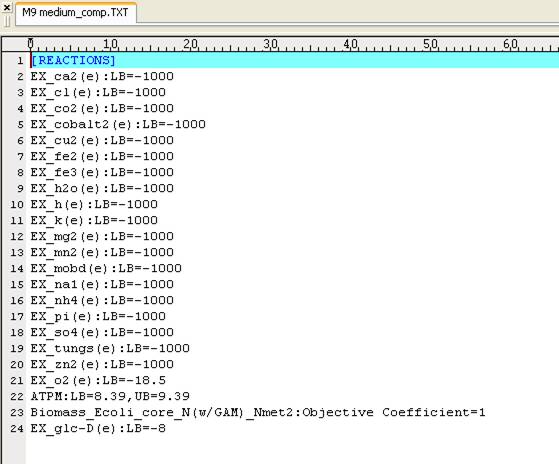
In silico minimal media for the
model iAF1260.
A complete
medium to simulate all extracellular metabolites can enter/exit the cell
freely.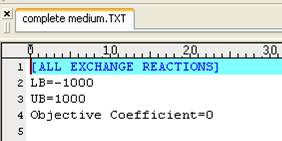
The
user-defined system boundaries and the objective are set in the reconstruction
model accordingly.
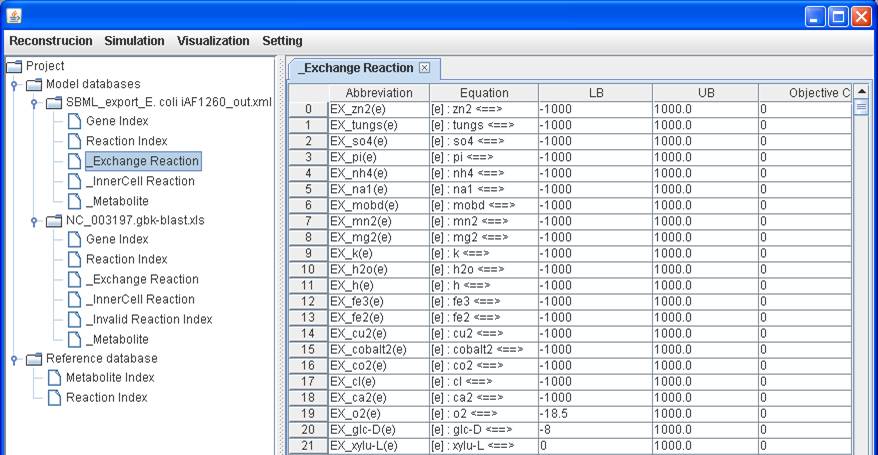
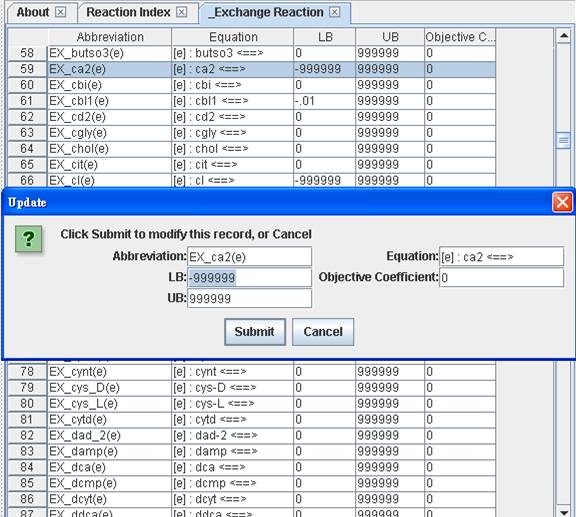
Or you can
simply right click on the reaction to update the lower bound (LB), upper bound
(UB) or objective coefficient.
You can
freely export or save a metabolic model in SBML format or in spreadsheet format
by right clicking on a model to Export model (.xml) or to Download
model (.xls). Such models generated by GEMSiRV are fully compatible to GEMSiRV
for later importing and simulation.
In addiction
to the metabolic models saved in SBML format, metabolic reconstructions can be
stored in spreadsheet format. The spreadsheet format can store the two-layer
relation for gene-protein and protein-reaction associations in network
reconstructions. We provide available reconstruction models (GPR) in http://sb.nhri.org.tw/GEMSiRV/en/Metabolic_Models
and demonstrate how we use GEMSiRV to reconstruct metabolic networks with GPR
relationships.
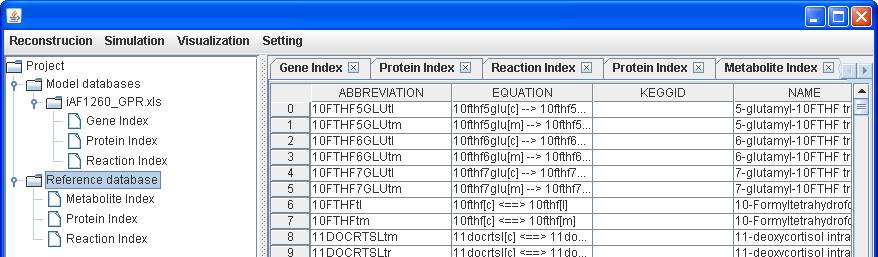
From
reconstruction to model
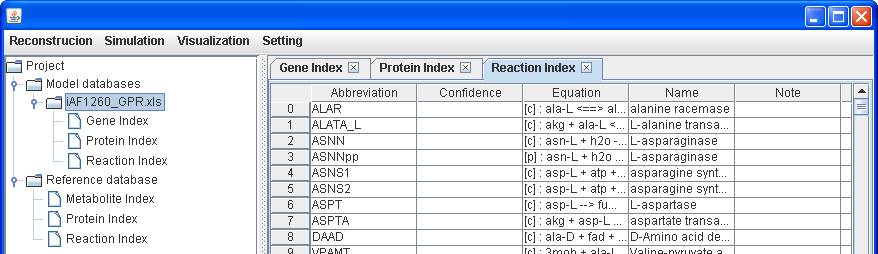
After
clicking on Reconstruction in the menu bar, right click on the Model databases to Import
spreadsheets (.xls) for importing the reconstruction file of
iAF1260_GPR.xls (download from http://sb.nhri.org.tw/GEMSiRV/en/Metabolic_Models).
This reconstruction contains three indices: Gene, Protein and Reaction Index.
Then right
click on Reference database to Import
database (.xls) for importing the reference database file Ref_BiGG_GPR.xls
which is provided in http://sb.nhri.org.tw/GEMSiRV/en/Reference_Databases.
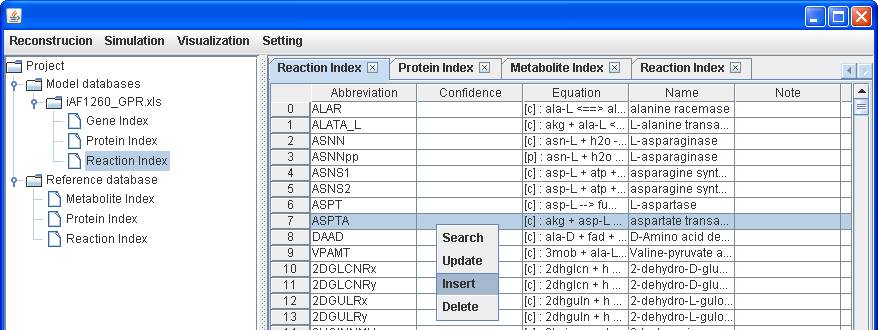
A biomass for
E. coli, Biomass_Ecoli_core_N (w/ GAM)-Nmet2, is available in the reference
database, you can add the reaction to the reconstruction by right clicking on
the main window of Reaction Index to Insert. After submitting the abbreviation
of reaction “Biomass_Ecoli_core_N (w/ GAM)-Nmet
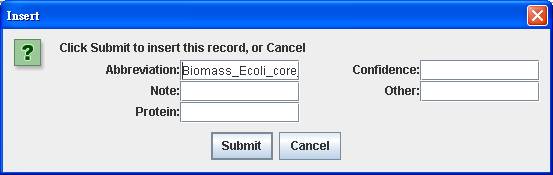
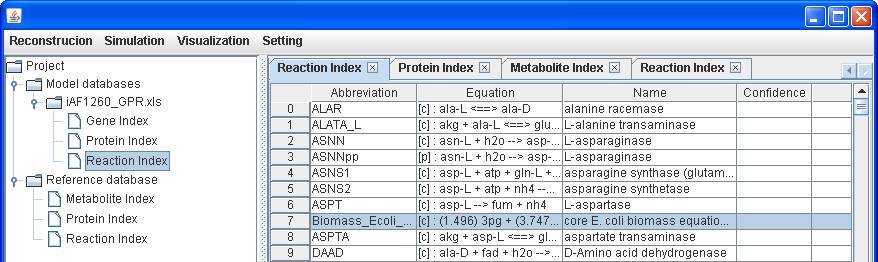
Likewise, you
can add a new reaction into the reference.
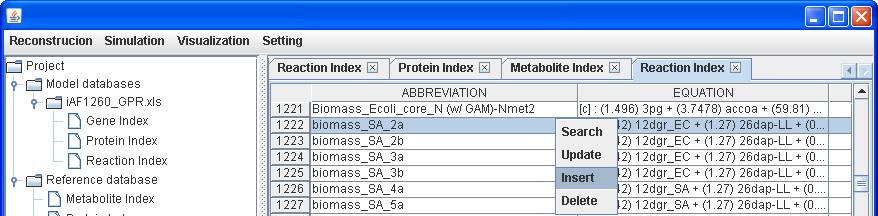
The reaction
of Ec_biomass_iAF1260_core_59p
Abbreviation:
Ec_biomass_iAF1260_core_59p
Equation:
(0.000223)
10fthf[c] + (0.000223) 2ohph[c] + (0.5137) ala-L[c] + (0.000223) amet[c] +
(0.2958) arg-L[c] + (0.2411) asn-L[c] + (0.2411) asp-L[c] + (59.984) atp[c] +
(0.004737) ca2[c] + (0.004737) cl[c] + (0.000576) coa[c] + (0.003158)
cobalt2[c] + (0.1335) ctp[c] + (0.003158) cu2[c] + (0.09158) cys-L[c] +
(0.02617) datp[c] + (0.02702) dctp[c] + (0.02702) dgtp[c] + (0.02617) dttp[c] +
(0.000223) fad[c] + (0.007106) fe2[c] + (0.007106) fe3[c] + (0.2632) gln-L[c] +
(0.2632) glu-L[c] + (0.6126) gly[c] + (0.2151) gtp[c] + (54.462) h2o[c] +
(0.09474) his-L[c] + (0.2905) ile-L[c] + (0.1776) k[c] + (0.01945)
kdo2lipid4[e] + (0.4505) leu-L[c] + (0.3432) lys-L[c] + (0.1537) met-L[c] +
(0.007895) mg2[c] + (0.000223) mlthf[c] + (0.003158) mn2[c] + (0.003158)
mobd[c] + (0.01389) murein5px4p[p] + (0.001831) nad[c] + (0.000447) nadp[c] +
(0.011843) nh4[c] + (0.04148) pe160[p] + (0.02233) pe160[c] + (0.02632)
pe161[c] + (0.04889) pe161[p] + (0.1759) phe-L[c] + (0.000223) pheme[c] +
(0.2211) pro-L[c] + (0.000223) pydx5p[c] + (0.000223) ribflv[c] + (0.2158)
ser-L[c] + (0.000223) sheme[c] + (0.003948) so4[c] + (0.000223) thf[c] +
(0.000223) thmpp[c] + (0.2537) thr-L[c] + (0.05684) trp-L[c] + (0.1379)
tyr-L[c] + (0.000055) udcpdp[c] + (0.1441) utp[c] + (0.4232) val-L[c] +
(0.003158) zn2[c] --> (59.81) adp[c] + (59.81) h[c] + (59.806) pi[c] +
(0.7739) ppi[c]
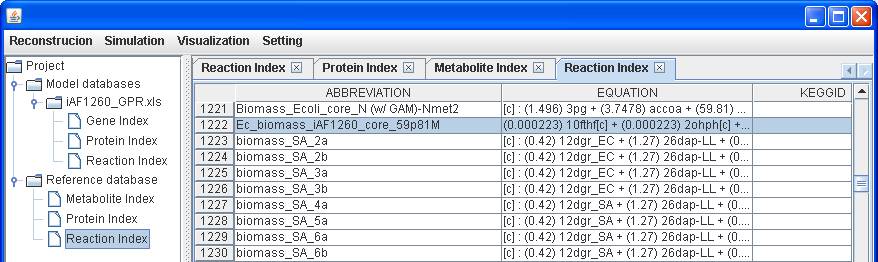
Right click
on a reconstruction to Generate simulation tables can convert the
reconstruction to a model. Then you can set the system boundaries for
simulation.
Draft
reconstruction and network refinement
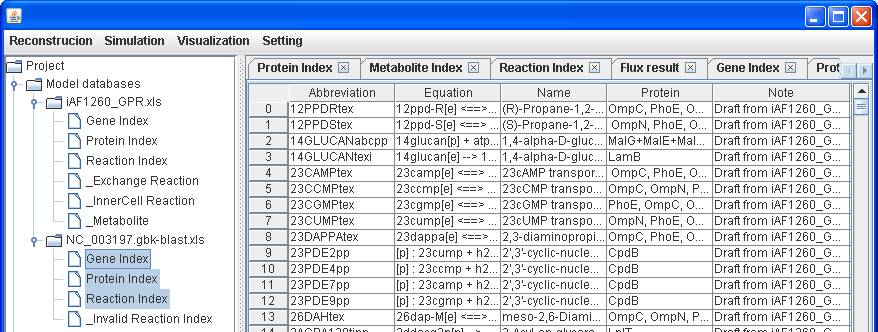
As described
previously, we can draft a reconstruction for a genetically related species
(e.g. Salmonella) with the existing E. coli model in GEMSiRV. Therefore, we
import the file NC_003197.gbk-blast.xls and draft a reconstruction with
reference to iAF1260_GPR.
Then we can
refine the draft reconstruction by adding metabolic reactions with
gene-protein-reaction associations, some existing reactions in the reference
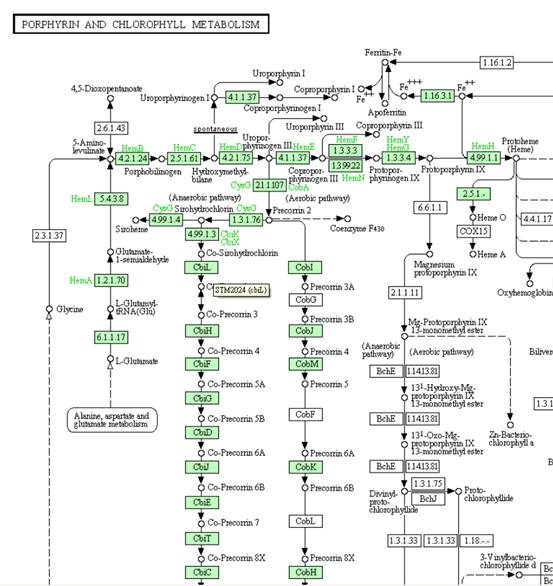
database can be conveyed to the reconstruction. For example, Salmonella is reported to be able to synthesize
cobalamin due to its metabolic genes (operon) STM2016-STM2035. Therefore, we
can manually add those associated reactions and proteins to the draft
reconstruction.
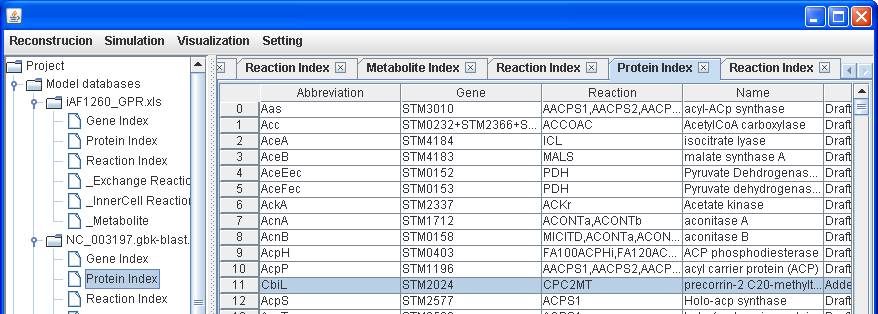
Reaction CPC2MT Name precorrin Equation [c]
: amet + copre2 --> ahcys + copre3 + h Locus Gene Protein Reaction CPC2MT STM2024 cbiL CbiL
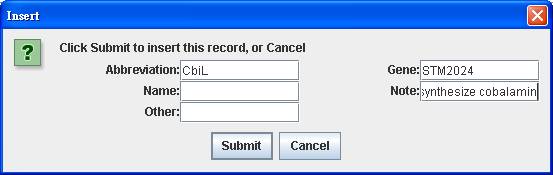
After clicking
into the Protein Index of NC_003197.gbk-blast.xls, right click on the main
window of protein index to insert the protein abbreviation CbiL, the associated
gene STM2024 and a note Added to synthesize cobalamin.
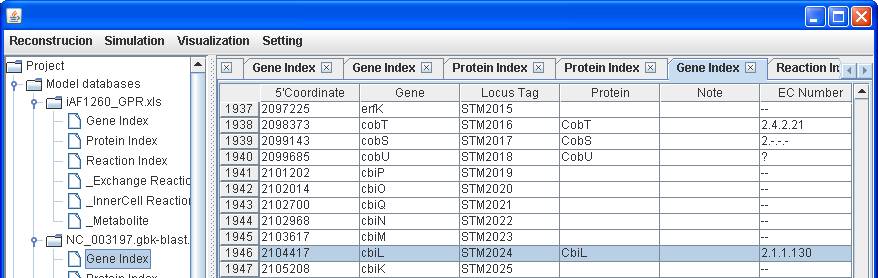
The
gene-protein association will be automatically brought into the Gene Index
table.
![]()
After
clicking into the Reaction Index of NC_003197.gbk-blast.xls, right click on the
main window of reaction index to insert the reaction abbreviation CPC2MT, the
associated protein CbiL, a note Added to synthesize cobalamin and the confidence
score 3 for genetic evidence.
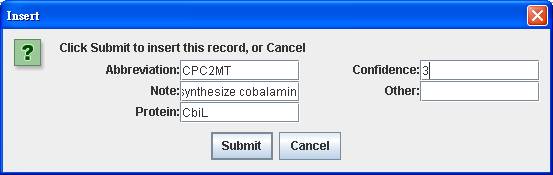
The reaction
information including name and equation will be automatically brought into the
Reaction Index table.
![]()
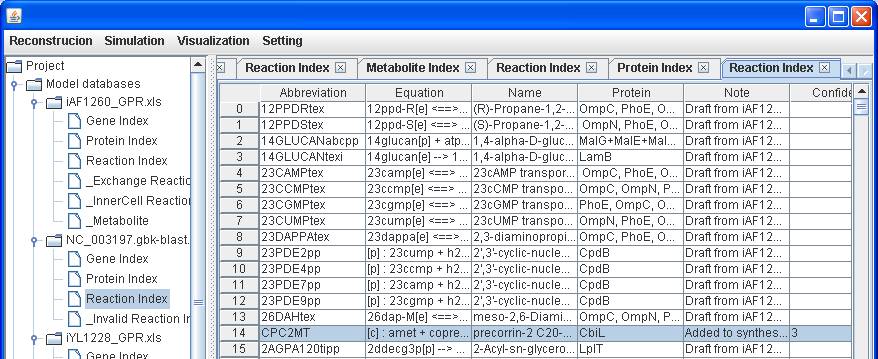
Likewise, the
protein-reaction association will be automatically brought into the Reaction
Index table.
![]()