Simulation
Before simulation, make sure you
have set the path of linear programming solver.
To download GNU Linear Programming Kit
(GLPK).
http://sourceforge.net/projects/winglpk/
(for windows) or
http://www.gnu.org/software/glpk/ (for Linux/Mac).
After extracting the file you downloaded (e.g.
winglpk-4.45.zip), please add the path of glpsol.exe to your Environment variables.
Open the Control Panel -> Click System -> Click
Advanced system setting -> Open Environment variables -> Edit Path -> Add
variable value ";the path where glpsol.exe locate" (e.g. ;D:\winglpk-4.45\w64)
Click on Simulation in the menu bar to choose which analysis you
want to perform.
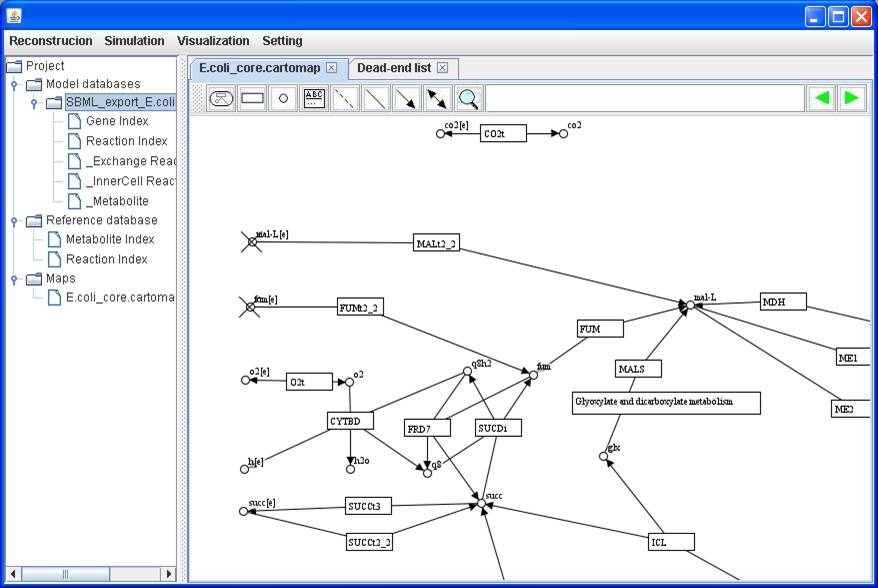
As a case study for demonstration of simulation, we import the E.coli textbook model which was exported from the BiGG into GEMSiRV and use a
customized map E.coli_core.cartomap for
visualization. You can find and download the model and the map from http://sb.nhri.org.tw/GEMSiRV/en/Metabolic_Models
and http://sb.nhri.org.tw/GEMSiRV/en/Metabolic_Maps,
respectively.
Dead-end metabolite identification
A network reconstruction is converted into a
mathematical model including a stoichiometric matrix
which describes the connectivity feature of the network and defined systems
boundaries before simulation. GEMSiRV can examine the
connectivity of all metabolites in a network for dead-end metabolite
identification and tag such metabolic dead ends with crosses in the map.
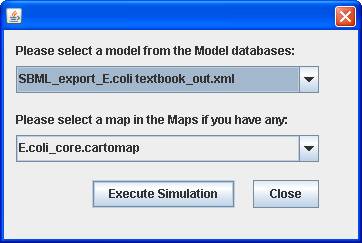
You can select a model and a map (if you have) to
perform the examination of network connectivity for dead-end metabolite
identification.
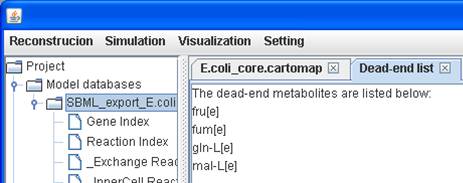
A dead-end
metabolite list is generated and those metabolites are tagged with crosses in
the map.
A list for dead-end metabolites:
A visualization
map with dead-end metabolites: