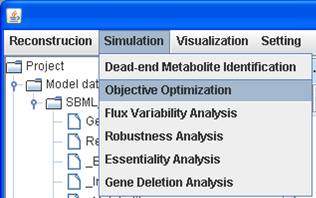
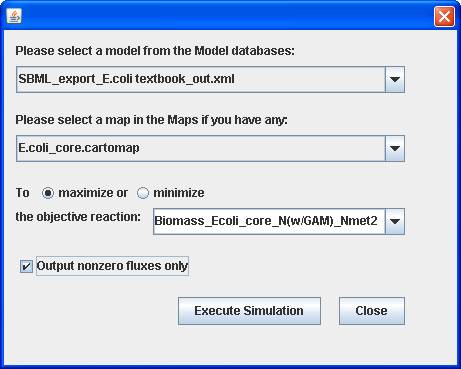
Objective optimization
With a set linear programming solver (e.g. glpk), GEMSiRV can be used to
simulate the imported metabolic network model. Please refer to http://sb.nhri.org.tw/GEMSiRV/en/Installation
for setting up GEMSiRV. Given proper constraints and
objective function, the flux results of all reactions in the model will be
estimated.
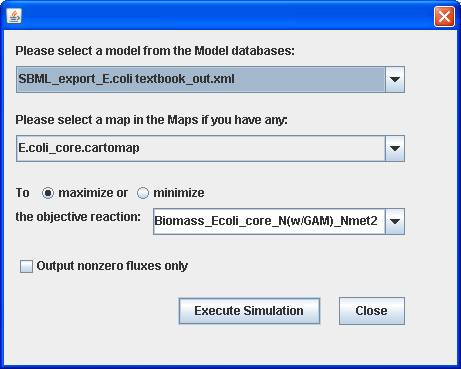
You can select a model and a map (if you have) for
objective optimization. The flux results can be visualized in the map.
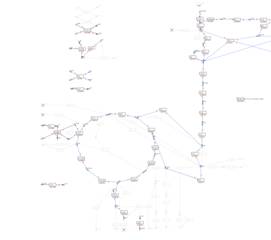
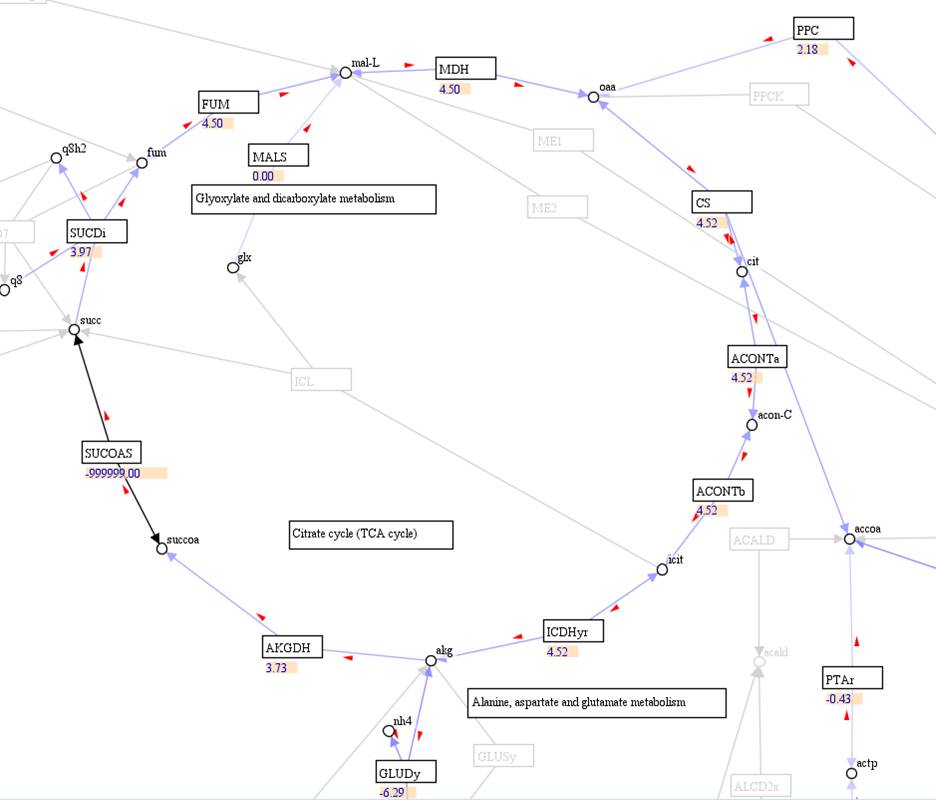
A visualization
map with reaction fluxes:
Scroll up on the map to zoom out:
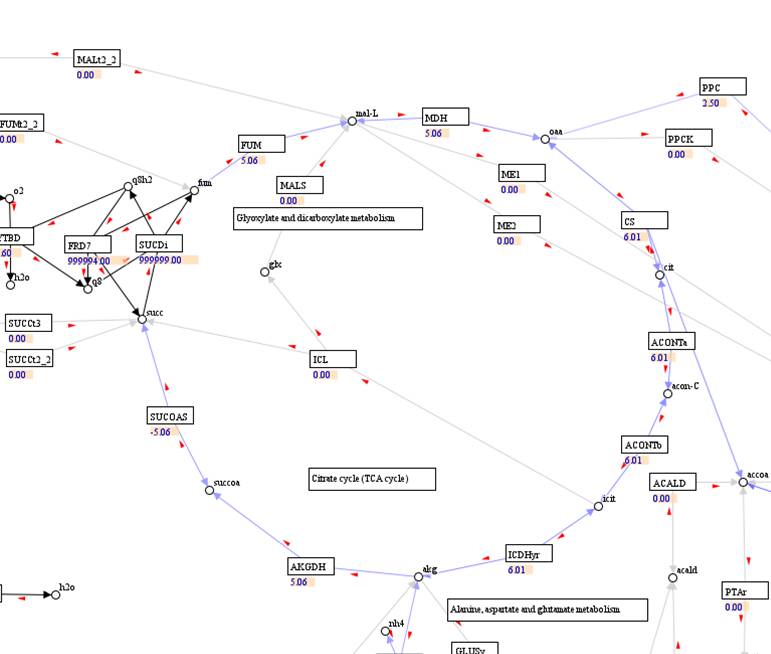
Flux result:
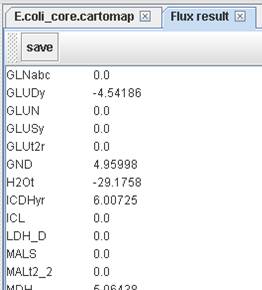
To check the
checkbox for outputting nonzero fluxes only.
Scroll up on the map to zoom out: