KEGG map loading
Click on Visualization in the menu bar to Load
KEGG maps by either Import KEGG map (.xml) or Retrieve KEGG map
depending on whether you have KGEE maps in hand.
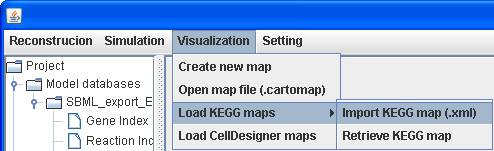
If not, you can click on Setting in the menu
bar to KEGG pathway Configure and set the link to where the KEGG pathway
maps can be retrieved, e.g. http://www.genome.jp/kegg-bin/download.
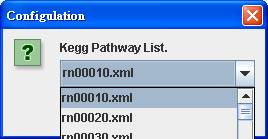
Then you can
retrieve KEGG pathway by choosing from the KEGG Pathway List.
The KEGG
pathway map of rn00010:
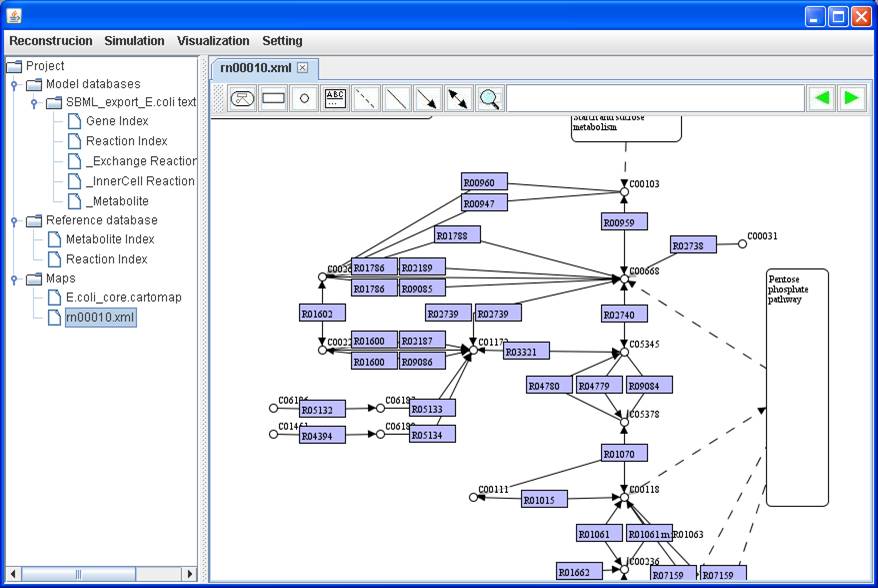
We set
rectangular nodes to represent reactions and define node name and node caption
for each reaction. We directly use the entry name and reaction in KEGG maps as
the node name and node caption respectively. Therefore, you can decide to Show
node name or Show node caption by right clicking on a map.
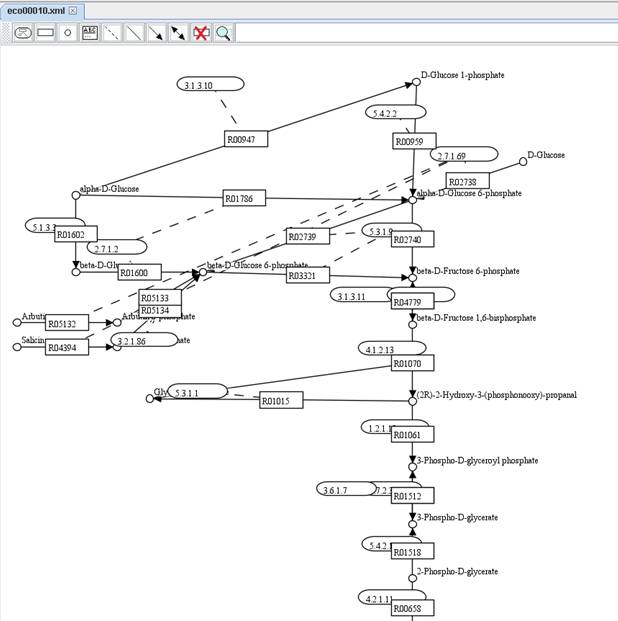
Content of KEGG pathway map (rn00010.xml):
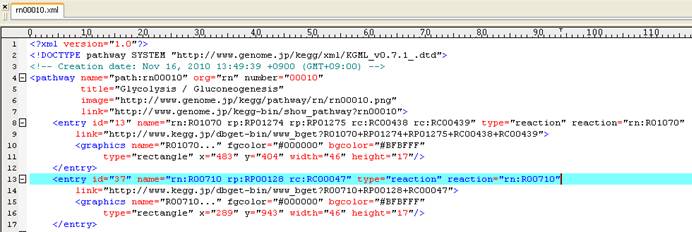
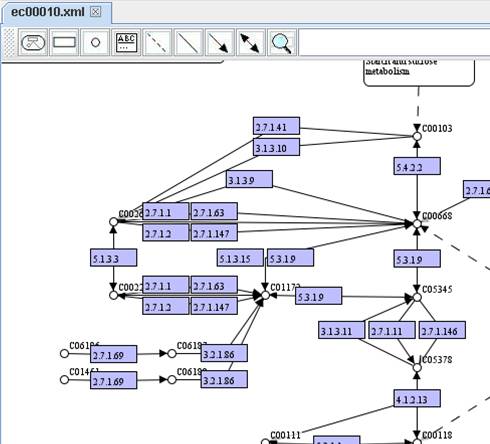
Content of
KEGG pathway map (ec00010.xml):
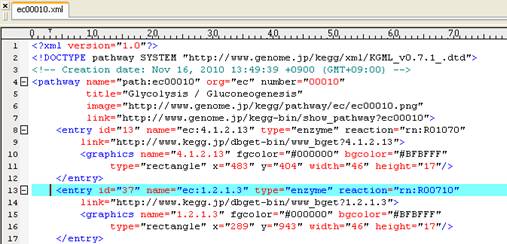
Show node
name:
Show
node caption:
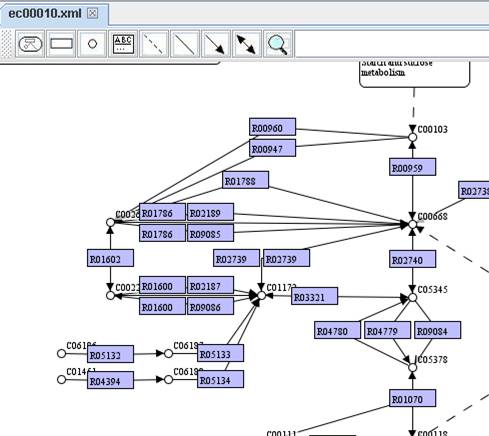
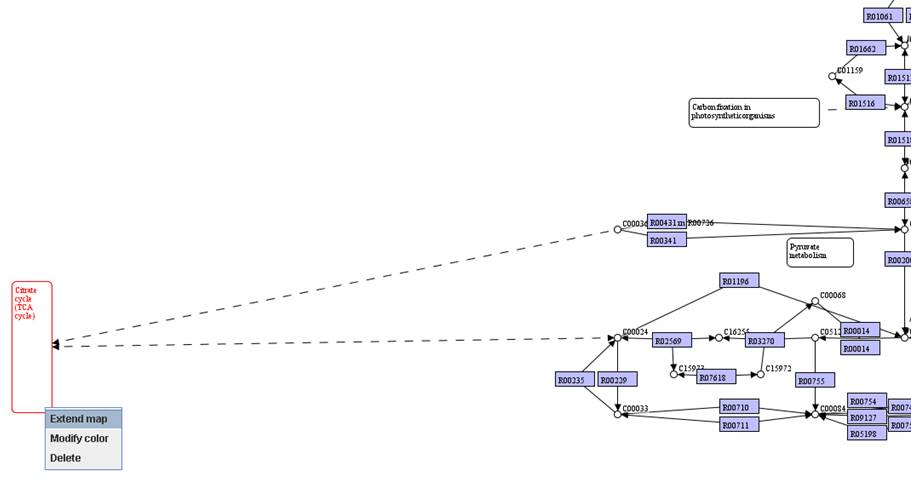
You can extend
other pathway maps in the map you have in the main network view window. A pathway
map is represented in a rounded rectangle. We can move the pathway that you
would like to extend to an empty region and right click on it to Extend map.
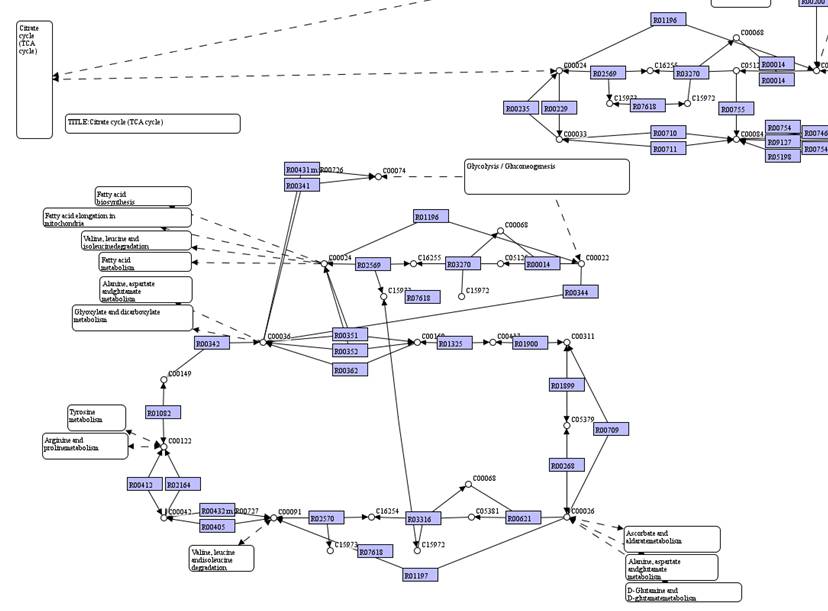
Map of
Citrate cycle (TCA cycle) is extended in the map:
You can hold
right-click button on the map and drag a rectangular region for selecting
groups of objects, then right click over the selected objects to delete them
all.
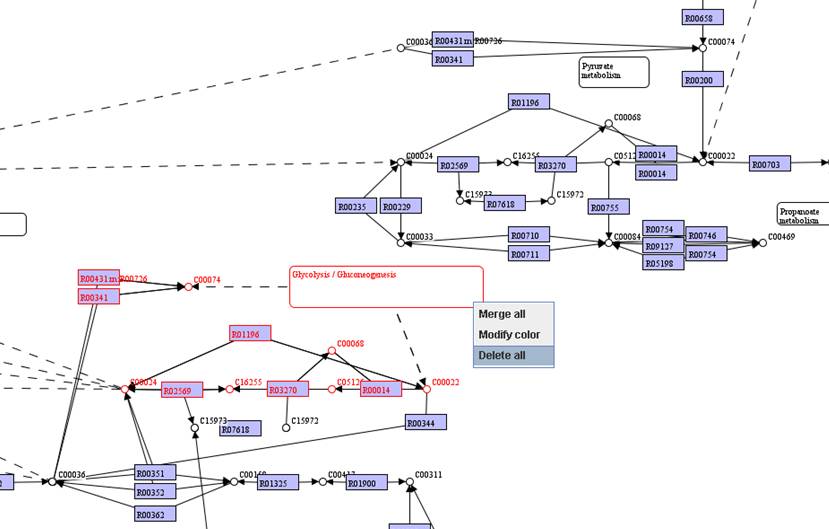
You can move
identical objects close to each other. Select the identical objects and then
right click on them to Merge all.
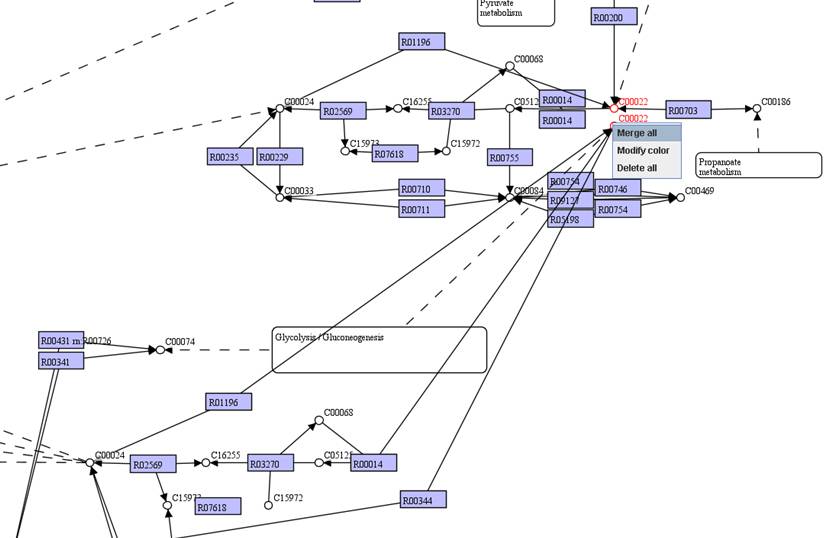
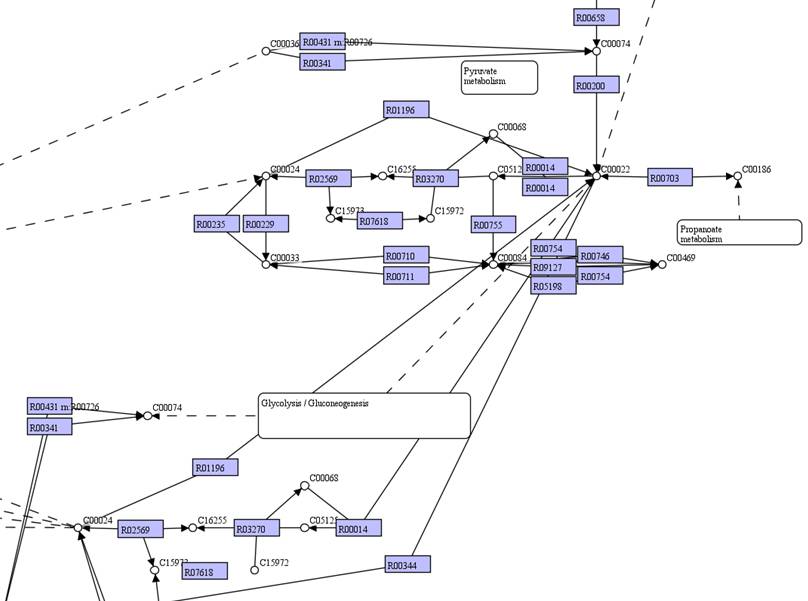
You can also
load SBML models compatible to CellDesigner (http://www.celldesigner.org/index.html)
to GEMSiRV. The SBML models for KEGG can be found and
downloaded in http://www.systems-biology.org/001/001.html.
You can click on Visualization in the menu bar to Load CellDesigner maps.
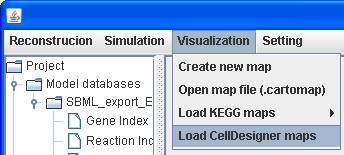
A SBML file
eco00010.xml provided in http://sb.nhri.org.tw/GEMSiRV/en/Metabolic_Maps
can be downloaded for demonstration.