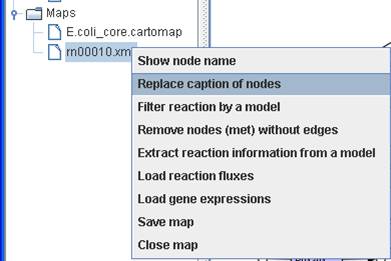
Map replacement
In order to ease the creation of customized maps, GEMSiRV provide a function in map replacement. You can
right click on a map to Replace caption of
nodes to convert the map to a customized map.
For example,
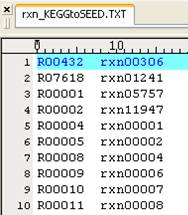
we replace a KEGG map (e.g. rn00010.xml) to a Model SEED-based map by providing
two separate lists for metabolite and reaction mapping. The KEEG to Model SEED
mapping lists can be found and downloaded in http://sb.nhri.org.tw/GEMSiRV/en/Manual.
Therefore,
some nodes of metabolite and reaction can be replaced to form a Model
SEED-based map.
Please
remember to save a map before closing it.
You can open
a map saved in cartomap format.
In order to
create a useful map for visualization, an interactive function between model
reconstruction and map visualization is implemented in GEMSiRV.
For
demonstration, we import a Model SEED model Acinetobacter
sp. ADP1 (Opt 62977.3.xml).
You can
filter reactions by comparing with the metabolic model you select and you can
get the reaction lists for reactions not existing or existing in the model.
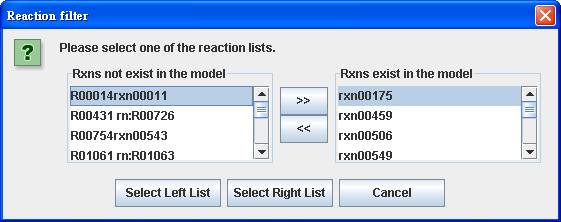
We select and
delete the left list of reactions for creating a model-specific map.
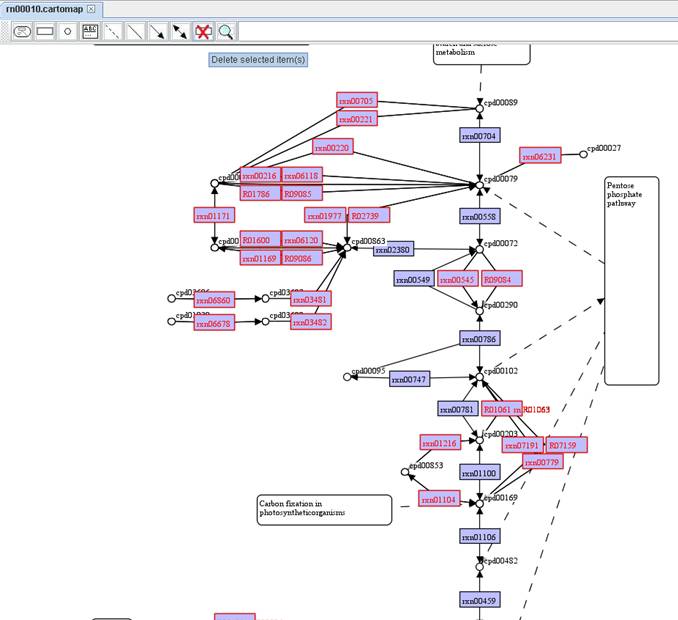
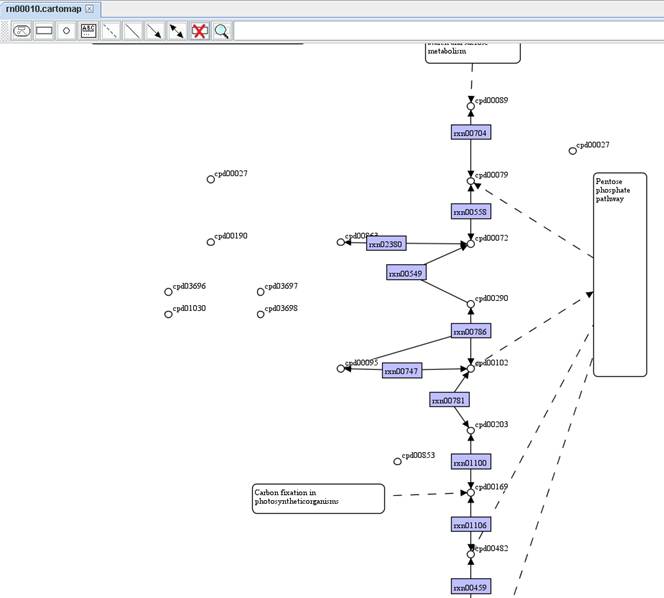
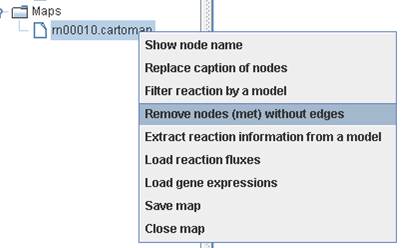
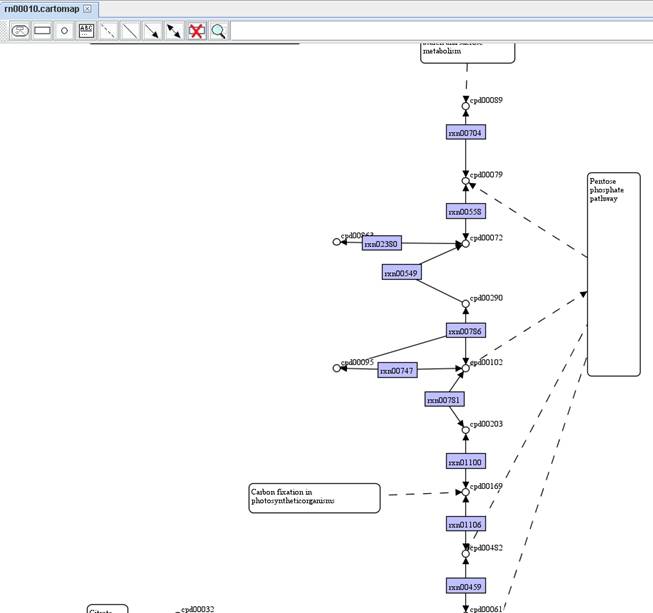
You can
remove those nodes of metabolite without linking to reaction by right clicking
on the map to Remove nodes (met) without
edges.
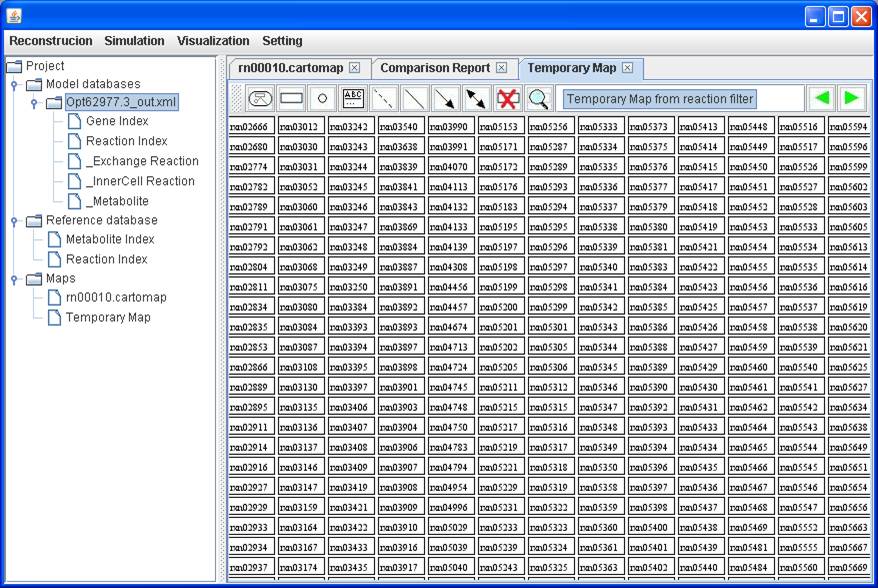
You can also
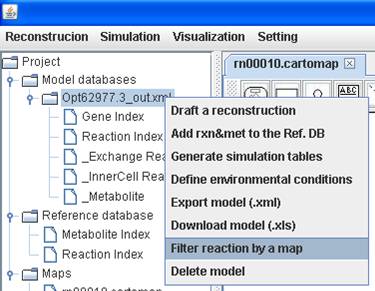
filter reactions in a metabolic model by comparing with a map. Right click on
the model to Filter reaction by a map and choose a map you want to
compare with. Then you can get a comparison report as well as a temporary map
including those reactions not present in the map you chosen.
A comparison report showing what reactions are present in the model only,
in the map only, and in the both.
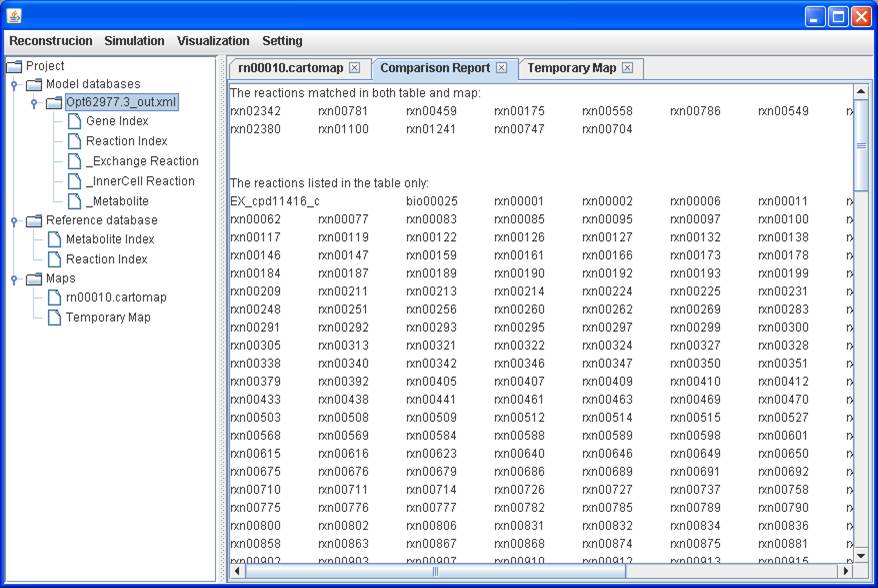
A temporary
map including those reactions in the model but not in the map
You can save
the temporary map and add it into the map you are working with.
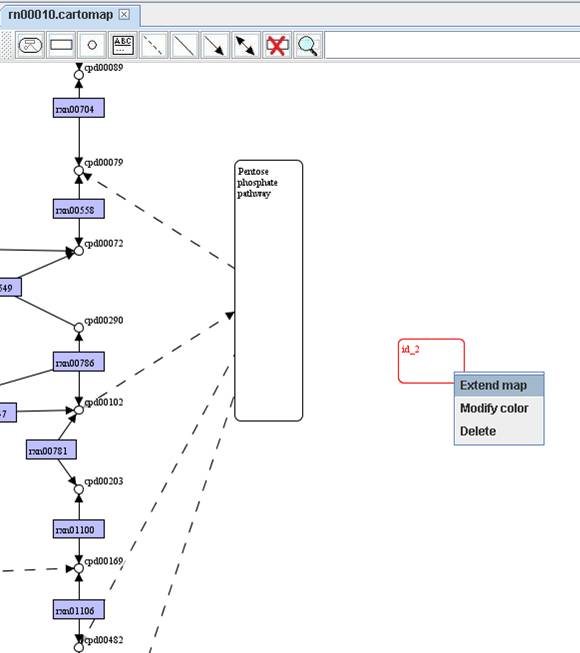
Add a map by
clicking Add a map in the toolbar and dropping in an empty region of the
map and extend the map by right clicking on the added map to Extend map.