A software platform for GEnome-scale Metabolic models Simulation, Reconstruction and Visualization.
This work has been published in Bioinformatics (Pubmed). If you have any question, please do not hesitate to contact the author by emailing to sysbio@nhri.org.tw.
- Start GEMSiRV An example model (E. coli textbook model) and its map can be directly imported by click on the Help to Import an Example.
Genome-scale metabolic network models have become an indispensable part of the increasingly important field of systems biology.
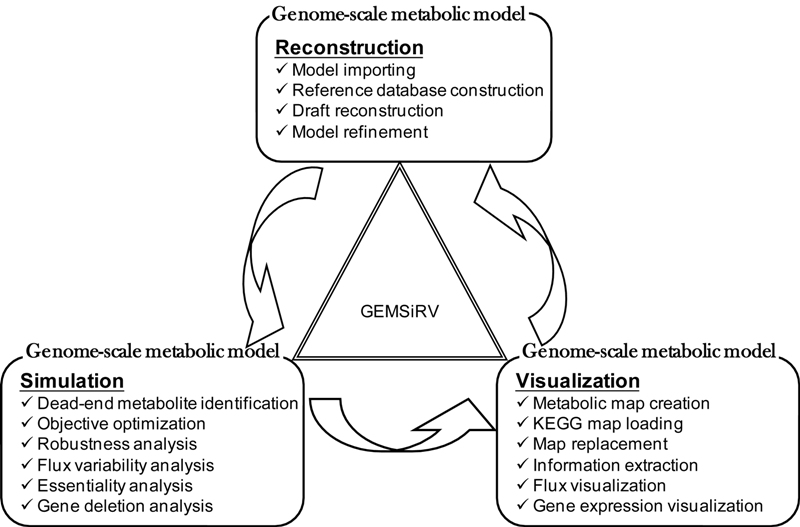
Metabolic systems biology studies usually include three major components – network model construction, objective- and experiment-guided model editing and visualization, and simulation studies based mainly on flux balance analyses.
Here we present a software platform, GEMSiRV, to provide functionalities of easy metabolic network drafting and editing, amenable network visualization for experimental data integration, and flux balance analysis tools for simulation studies.
GEMSiRV is an open-source software for building users' metabolic systems biology project, it provides interactive features in model management, simulation, visualization and integration of omics data.
Furthermore, all of the GEMSiRV-generated metabolic models and analysis results, including projects in progress, can be easily exchanged in the research community. GEMSiRV is a powerful integrative resource that may facilitate the development of systems biology studies.
A schematic overview of the GEMSiRV outlines the key features implemented in the GEMSiRV.