Completing bacterial genome assemblies: strategy and performance comparisons
Determining the genomic sequences of microorganisms is the basis and prerequisite for understanding their biology and functional characterization. While the advent of low-cost, extremely high-throughput second-generation sequencing technologies and the parallel development of assembly algorithms have generated rapid and cost-effective genome assemblies, such assemblies are often unfinished, fragmented draft genomes as a result of the short read lengths and long repeats present in multiple copies. Third-generation, PacBio sequencing technologies address this problem by greatly increasing read length. Hybrid and non-hybrid approaches, have therefore been proposed to utilize the PacBio long reads that can span many thousands of bases to facilitate the assembly of complete microbial genomes. However, standardized procedures that aim at evaluating and comparing these approaches are currently insufficient.
We therefore provide a comprehensive comparison by collecting datasets for the comparative assessment. In addition to offering explicit and useful recommendations to practitioners, this study aims to aid in the design of a paradigm positioned to complete bacterial genome assembly.
Following a special methodology proposed by ALLPATHS-LG, the algorithm is supplied with three pre-prepared libraries—fragment, jump and long reads. ALLPATHS-LG is subsequently able to complete bacterial genomes as the sequencing coverage of fragment library is controlled at 100X. Although other hybrid approaches (including PacBio corrected reads pipeline, SPAdes, SSPACE-LongRead) could greatly improve continuity over the assembly produced by second-generation sequencing reads, we have demonstrated that such a hybrid approach is not efficient way to complete bacterial genomes. Both non-hybrid approaches—hierarchical genome-assembly process and PacBio corrected reads pipeline via self-correction—are able to produce complete genomes provided that the third generation sequencing reads are adequately long and complete.
Datasets employed in this study
We have used ALLPATHS-LG (v44837) and SPAdes(v3.1.0) to assemble three bacterial genomes: E. coli, R. sphaeroides, and S. pneumoniae. The sequencing reads for these three genome assemblies are summarized in the following table (D1-D3).
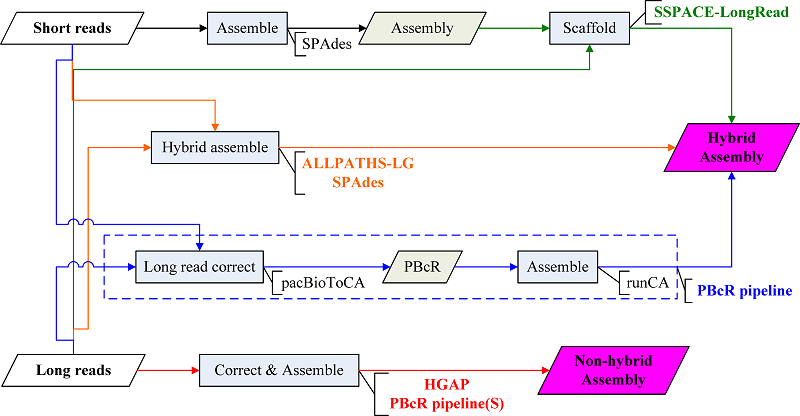
We have conducted PBcR pipeline proposed by Koren et al.(ref) to correct long reads (D5) with short reads (D4) (by PacBioToCA), then to de novo assemble the corrected long reads (by runCA) for E. coli genome reconstruction. We firstly investigated the effect of sequencing depths on assembly (Read Depths), then set genome size in running pacBioToCA, finally we tried different Celera Assembler parameters for runCA. SPAdes 3.1 is able to directly hybrid assemble the combined dataset (D4+D5). In addition, a scaffolder, named SSPACE-LongRead (v1-1), was used to scaffold pre-assembled contigs constructed from short reads (D4) using long reads (D5).
We have conducted the both non-hybrid approaches: hierarchical genome-assembly process, HGAP(v2.0), and PBcR pipeline via self-correction (PBcR pipeline) to de novo assemble the PacBio long reads. The datasets used in the non-hybrid approach are composed of various SMRT cells, ranging from 4 to 17 XL-C2 SMRT cells (D5-D8), and a single SMRT cell gathered with PacBio RS II system and P4-C2 chemistry (D9).
| Data | Organism | Fragment | Jump | Long read | Reference |
| D1 | E. coli K-12 MG1655 | 2×101 bp, 180 bp insert (SRR447685) | 2×93 bp, 3000 bp insert (SRR401827 and SRR492488) | 1-3 Kbp (Ribeiro's ftpa) | NC_000913 |
| D2 | R. sphaeroides 2.4.1 | 2×101 bp, 180 bp insert (SRR125492) | 2×101 bp, 3000 bp insert (SRR388672) | 1-3Kbp (Ribeiro's ftpa) | NC_007488-90, NC_007493-94, NC_009007-08 |
| D3 | S. pneumoniae Tigr4 | 2×101 bp, 180 bp insert (SRR387335) | 2×93 bp, 3000 bp insert (SRR364158) | 1-3 Kbp (Ribeiro's ftpa) | NC_003028 |
| D4 | E. coli K-12 MG1655 | 2×151 bp, 300 bp insert (Illumina data websiteb) | NC_000913 | ||
| D5 | E. coli K-12 MG1655 | 10 Kbp, 17 SMRT cell (SRX255228c) | NC_000913 | ||
| D6 | E. coli K-12 MG1655 | 8-10 Kbp, 8 SMRT cells (SRX260475d) | NC_000913 | ||
| D7 | M. ruber DSM1279 | 8-10 Kbp, 4 SMRT cells (SRX260496d) | NC_013946 | ||
| D8 | P. heparinus DSM2366 | 8-10 Kbp, 7 SMRT cells (SRX260506d) | NC_013061 | ||
| D9 | E. coli K-12 | PacBio RS II System and P4-C2 chemistrye, 20 Kbp library, 1 SMRT cell | NC_000913 |
a Long reads were downloaded from ftp://ftp.broadinstitute.org/pub/papers/assembly/Ribeiro2012/data
b Paired reads were provided in http://www.illumina.com/systems/miseq/scientific_data.ilmn
c PacBio HDF5 files were requested from NCBI Sequence Read Archive (SRA).
d PacBio HDF5 files were downloaded from http://files.pacb.com/software/hgap/index.html
e PacBio HDF5 files were downloaded from https://github.com/PacificBiosciences/DevNet/wiki/E.-coli-20kb-Size-Selected-Library-with-P4-C2
Workflow
SSPACE-LongRead is a scaffolder using single molecule long reads to upgrade pre-assembled contigs constructed from short reads. ALLPATHS-LG and SPAdes are hybrid assemblers which take short reads and long reads as inputs. PBcR pipeline uses short reads to correct long reads by pacBioToCA, and then assembles corrected long reads (PBcR) by Celera assembler (runCA). Hierarchical genome-assembly process (HGAP) and PBcR pipeline via self-correction (PBcR pipeline(S)) take long reads as input to produce non-hybrid assembly.