CISA
Contig Integrator for Sequence Assembly
Recently, technological advances have dramatically improved throughput and quality of next-generation sequencing (NGS), and, in parallel with these improvements, numerous algorithms have been proposed for de novo sequence assembly. Compared to the traditional Sanger sequencing technology, NGS technologies offer several distinct features, such as large volumes of reads and concise length. In order to tackle the sequence assembly problem from a collection of short-sequencing reads of randomly sampled fragments, two types of algorithm, the overlap-layout-consensus approach and the de Bruijn graph, are commonly utilized (Paszkiewicz and Studholme 2010; Aerts, Narzisi et al. 2011). Although assemblers are generally based on a small number of algorithms, they differ from each other in terms of dealing with errors, inconsistencies, and ambiguities. Moreover, no individual assembler guarantees the best assembly for diverse species. Therefore, the use of various parameter settings and assemblers in an iterative manner to produce an improved draft assembly is inevitable. Nevertheless, few efforts have been made to integrate various assemblies into a draft that is marked by superior contiguity and accuracy. To the best of our knowledge, five tools, including GAA (Yao, Ye et al. 2012), GAM (Casagrande, Del Fabbro et al. 2009), MAIA (Nijkamp, Winterbach et al. 2010), minimus2 (Sommer, Delcher et al. 2007) and Reconciliator (Zimin, Smith et al. 2008), have been published for merging assemblies. However, GAM and Reconciliator require the original reads and complicated prerequisites; we therefore compared our proposed method with GAA, MAIA and minimus2.
The quality of genome assemblies is usually evaluated by means of their contiguity and the accuracy of contigs or scaffolds. Contiguity is a straightforward measurement achieved by calculating the N50 length or the number of contigs/scaffolds, without mandating reference genome information to be implemented in the algorithm. On the other hand, the accuracy of an assembly can be assessed based on alignment with a complete reference genome (Darling, Tritt et al. 2011). However, there is an inevitable trade-off between contiguity and accuracy in such scenarios. Specifically, an assembler attempting to maximize contiguity might do so at the expense of a less accurate assembly, and vice versa. Since an individual assembler has its own features in addressing the reconstruction of a DNA sequence, can we take advantage of all assemblies to generate an integrated set of contigs?
In this study, state-of-the-art assemblers were used to generate different sets of contigs for bacterial genomes. We have developed and built a Contig Integrator for Sequence Assembly (CISA) to integrate the sets of contigs from different assemblers, and have evaluated the quality of our integrated assembly. Compared with the assemblies generated by each individual assembler and assembly integrator, the hybrid assembly integrated by CISA exhibits superior contiguity and accuracy.
This work has been published in PLoS ONE (March 28, 2013).
News !!!
[Nev 25, 2014] We have built a Virtual Box (MyPro) tailored for prokaryotic genome assembly and annotation. Please visit http://sourceforge.net/projects/sb2nhri/files/MyPro/ for details.
[Aug 28, 2013] We have added frequently asked questions in Instruction#Troubleshooting.
[Aug 01, 2013] We have used CISA to integrate two additional examples (B. cereus and R. sphaeroides) and evaluated by QUAST.
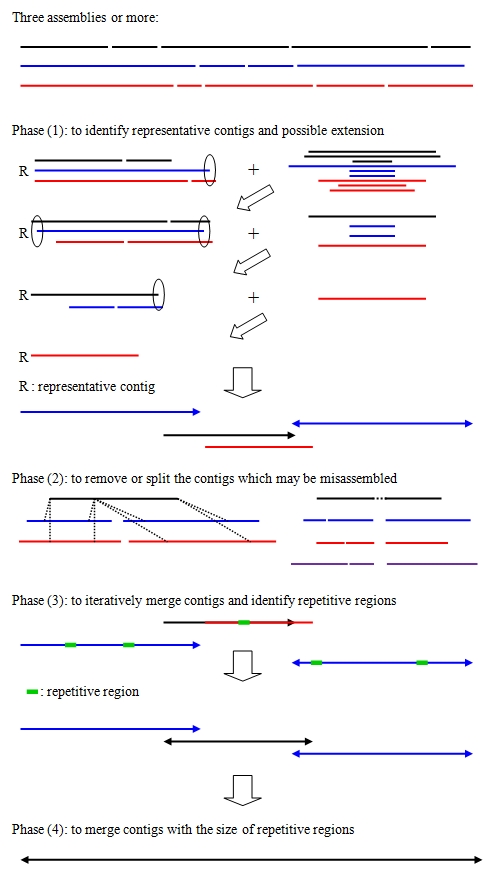
A schematic overview of CISA
CISA basically consists of four major phases:
- (1) identification of the representative contigs and possible extensions
- (2) removal and splitting of the contigs that may be misassembled, and clipping of uncertain regions that are located at the extremities of the contigs
- (3) iterative merging of the contigs with a minimum 30% overlap and estimating the maximal size of repetitive regions
- (4) merging of the contigs based on the size of repetitive regions
CISA performs these four phases and generates four folders accordingly. The detailed description about CISA output can be seen in Description.
In addition, the detailed information about how to use CISA can be seen in Instruction.
We have used three bacterial genome assemblies to test CISA, and compared the results with the assemblies generated by various assemblers or merged tools.