CISA
Contig Integrator for Sequence Assembly
Recently, technological advances have dramatically improved throughput and quality of next-generation sequencing (NGS), and in parallel with the improvements, many algorithms have been proposed for de novo sequence assembly. Compared to the traditional Sanger sequencing technology, the NGS technologies offer several distinct features, such as large volumes of reads and short length. In order to tackle the sequence assembly problem from a collection of short sequencing reads of randomly sampled fragments, two types of algorithms–overlap-layout-consensus approach and the de Bruijn graph–are commonly utilized. Albeit the assembler are mainly based on the small number of algorithms, they differ from each in terms of dealing with errors, inconsistencies and ambiguities. Moreover, no individual assembler guarantees the best assembly of diverse species. Performing different parameter settings or different assemblers in an iterative manner to generate a draft assembly is inevitable. Nevertheless, few efforts have been made to integrate the various assemblies into a better draft which possess superior quality in both contiguity and accuracy
The qualities of genome assemblies are usually evaluated by their contiguity and the accuracy of contigs or scaffolds. The contiguity is a straightforward measurement by calculating the N50 length or the number of contigs/scaffolds, no need of the real genome. On the other hand, the accuracy of an assembly can be assessed based on alignment to a complete reference genome (Darling, et al., 2011). However, there is a transparent trade-off between contiguity and accuracy. In other words, an assembler trying to maximize the contiguity might provide a less accurate assembly, and vice versa. Since each assembler has its own features in addressing the reconstruction of a DNA sequence, can we take the advantages of all the assemblies to generate an integrated set of contigs?
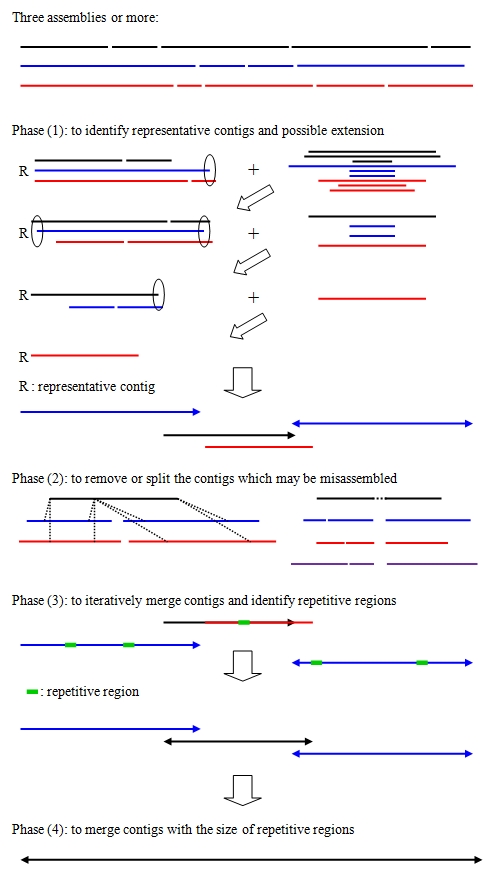
In this study, utilizing state-of-the-art assemblers to generate different sets of contigs for bacterial genomes is conducted. We have developed and built a Contig Integrator for Sequence Assembly (CISA) to integrate the sets of contigs from different assemblers and evaluated the quality of our integrated assembly. Comparing with the assembly generated by each individual assembler, the assembly sequences integrated by CISA has superior contiguity.
A schematic overview of CISA
Bascially