E. coli 36 bp
Escherichia coli K12 MG1655. The E. coli MG1655 consists of a circular chromosome of 4,639,675 bp in length.
Read source
- The illuminia read data of E. coli (Paired-end 36 bp sequencing library with 200 bp inserts) were downloaded from Sequence Read Archive (SRA). More than 20.8 M reads
Sequence assembly
- Set1 (Different Assemblers)
| Software | Version | Parameters | Download |
| ABySS | 1.3.0 | k=31 | Abyss |
| Velvet | 1.1.04 | k=29 ins_length=215 cov_cutoff=12 exp_cov=24 min_contig_lgth=100 scaffolding=no | Velvet |
| Edena | 3 | m=30 | Edena |
| SOAPdenovo | 1.05 | K=29 M=3 | SOAPdenovo |
| CLC | 4.7.2 | insert_size_range=194,236 minimum_contig_length=100 | CLC |
Merged File: Set1_Contig
- The above assemblers together with the parameter setting have been selected for de novo assembling of E. coli. After assembly, we discarded contigs with less than 100bp and evaluated the accuracy of the assemblies based on the Mauve Assembly Matrices (the results are shown below). In this set of data, we have different sequence assemblies, each was generated by the different assembler. Because the same assembler performs differently over varying parameter settings, such as kmer, we have tried different parameters for Abyss and SOAPdenovo in the following sets.
- Set2 (Different parameters for Abyss - the assembler provides the lowest number of contigs in Set1)
| Abyss parameter | Download |
| k=29 | Abyss_k29 |
| k=31 | Abyss_k31 |
| k=33 | Abyss_k33 |
Merged File: Set2_Contig
- Set3 (Different parameters for SOAPdenovo - the assembler provides the largest number of contigs in Set1)
| SOAPdenovo parameter | Download |
| k=29 | SOAP_k29 |
| k=31 | SOAP_k31 |
| k=33 | SOAP_k33 |
Merged File: Set3_Contig
- Scaffold the contigs using SSPACE
| Download |
| CISA+SSPACE |
| ABySS+SSPACE |
| Minimus2+SSPACE |
Contig integrator
- CISA
| Input | Download |
| Set1 | CISA_Set1 |
| Set2 | CISA_Set2 |
| Set3 | CISA_Set3 |
| Set2+Set3 | CISA_Set2_3 |
| Set1+2+3+2_3 | CISA_Set1+2+3+2_3 |
The input file for CISA is an assembled set of contigs, e.g., the set1 contains the contigs obtained from Abyss, CLC, Edena, SOAPdenovo, and Velvet.
The integrated contigs generated by CISA can be directly downloaded via the Download link.
- MAIA
maia('./assembly_list.txt','./NC_000913.fna')
assembly_list.txt:
Edena ./Edena_contigs.fa 2 SOAP ./SOAP_contigs.fa 2 Velvet ./Velvet.fa 2 CLC ./CLC.fa 2 Abyss ./Abyss_contigs.fa 3
A file named maia_assembly.fa was generated, containing a draft genome of 4640055 bp with 126761 uncalled bases (Ns). We split the genome into contigs (at >10Ns) and got the set of contigs maia.fa.
- minimus2
| Input | Download |
| Set1 | minimus2_Set1(A,C,E,S,V) |
| Set1 | minimus2_Set1(S,C,V,E,A) |
Since minimus2 can only merge two sequence sets at a time, we used minimus2 to integrate the set1 in alphabetical order (i.e., ACESV) and in descending order of contig number (i.e., SCEVA).
- GAA
After formatting the contig names in the format of >Contig1.1, we conducted GAA to integrate the assembly of Abyss with the assembly of Edena by using the following script and got the set of contigs gaa2.fa.
perl gaa.pl -t ../Abyss_contigs_1.fa -q ../CLC_1.fa -o AC
Similarly, we can iteratively integrate the output assembly with another assembly to get the set of contigs gaa5.fa (A+C+E+S+V).
Evaluation
- Benchmark genome
- Evaluate by Mauve Assembly Metrics
- Score with Mauve metrics:
Set1
| Name | NumContigs | NumAssemblyBases | DCJ_Distance | NumMisCalled | NumUnCalled | NumGapsRef | NumGapsAssembly | TotalBasesMissed | %Missed | ExtraBases | %Extra | BrokenCDS | IntactCDS | ContigN50 | ContigN90 | MaxContigLength | Blast_IntactCDS | Units(>200) | N50 | cor.Units | cor.N50 | Errors,(Indel>=5,Inv,Rel) |
| Abyss | 133 | 4626205 | 108 | 334 | 69 | 123 | 119 | 57847 | 1.2468 | 29424 | 0.636 | 57 | 4263 | 96157 | 26096 | 222425 | 4257 | 108 | 96511 | 116 | 92933 | 8,(6,0,2) |
| CLC | 379 | 4546926 | 304 | 100 | 0 | 288 | 287 | 130550 | 2.8138 | 3405 | 0.0749 | 62 | 4258 | 29767 | 8447 | 107342 | 4233 | 288 | 28450 | 290 | 28036 | 2,(0,1,1) |
| Edena | 211 | 4569446 | 154 | 17 | 0 | 129 | 125 | 86780 | 1.8704 | 2078 | 0.0455 | 66 | 4254 | 54405 | 13642 | 186686 | 4204 | 182 | 54405 | 186 | 52796 | 4,(2,1,1) |
| SOAPdenovo | 553 | 4547211 | 475 | 36 | 0 | 461 | 412 | 124407 | 2.6814 | 6972 | 0.1533 | 100 | 4220 | 17902 | 5384 | 103369 | 4146 | 450 | 17892 | 451 | 17892 | 1,(0,0,1) |
| Velvet | 283 | 4550675 | 207 | 138 | 0 | 208 | 203 | 116542 | 2.5119 | 2783 | 0.0612 | 74 | 4246 | 52474 | 12537 | 166094 | 4204 | 217 | 52474 | 224 | 49022 | 8,(5,0,3) |
| CISA_Set1 | 72 | 4627549 | 70 | 241 | 50 | 91 | 92 | 49487 | 1.0666 | 32028 | 0.6921 | 44 | 4276 | 119107 | 32288 | 312018 | 4290 | 69 | 135136 | 83 | 113511 | 14,(7,1,6) |
| MAIA | 110 | 4513348 | 96 | 82 | 54 | 100 | 95 | 129936 | 2.8005 | 1090 | 0.0242 | 48 | 4272 | 112717 | 30950 | 312145 | 4222 | 95 | 126075 | 97 | 107674 | 5,(2,0,3) |
| minimus2(A,C,E,S,V) | 74 | 4608653 | 68 | 285 | 0 | 97 | 78 | 76881 | 1.657 | 35464 | 0.7695 | 50 | 4270 | 126075 | 34542 | 417704 | 4268 | 73 | 134584 | 83 | 113511 | 10,(7,1,2) |
| minimus2(S,C,V,E,A) | 69 | 4215087 | 69 | 214 | 249 | 90 | 78 | 548181 | 11.8151 | 113137 | 2.6841 | 51 | 4269 | 119108 | 35441 | 312145 | 3869 | 69 | 115198 | 79 | 105796 | 10,(5,2,3) |
| GAA (Abyss,Edena) | 133 | 4637982 | 102 | 328 | 93 | 118 | 112 | 54835 | 1.1819 | 28888 | 0.6229 | 57 | 4263 | 96157 | 26096 | 222425 | 4267 | 108 | 96511 | 115 | 92933 | 8,(6,0,2) |
| GAA (A,C,E,S,V) | 138 | 4639673 | 103 | 305 | 93 | 119 | 113 | 54254 | 1.1693 | 29292 | 0.6311 | 57 | 4263 | 96157 | 26096 | 222425 | 4267 | 108 | 96511 | 115 | 92933 | 8,(6,0,2) |
- CISA integrates the five assemblies (Abyss, CLC, Edena, SOAPdenovo and Velvet from the outer to the inner) and sequentially generates the processed contigs after phase (1), (2), (3) and (4):
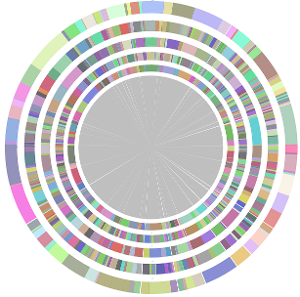
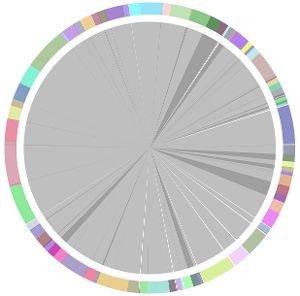
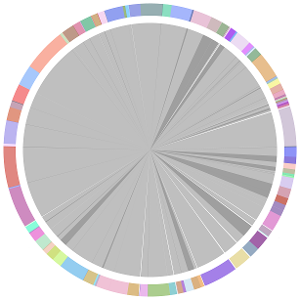
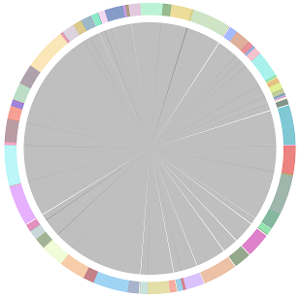
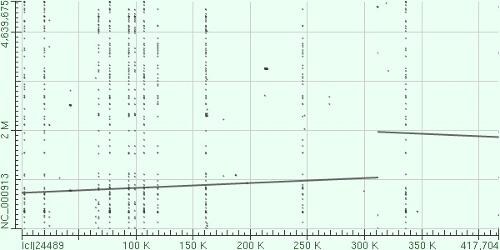
- We have visually inspected the assemblies against the reference genome (NC_000913) by using graphic representations, e.g. dot plots. Therefore, we knew that the largest contig generated by minimus2(A,C,E,S,V) was misassembled. Moreover, we found that the merging order dramatically influences the result of minimus2.
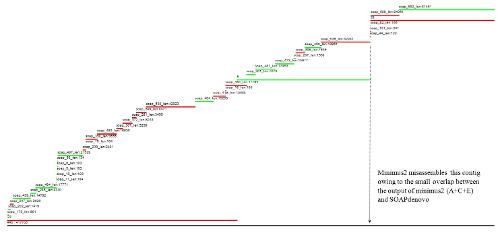
- Minimus2 firstly merges SOAPdenovo (the inner) with CLC (the outer), then merges the output (the inner) with Velvet (the outer) in the second run, merges the output (the inner) with Edena (the outer) in the third run, finally merges the output (the inner) with Abyss (the outer) in the fourth run and generates the integrated contigs.
Set2-Set3
| Name | NumContigs | NumAssemblyBases | DCJ_Distance | NumMisCalled | NumUnCalled | NumGapsRef | NumGapsAssembly | TotalBasesMissed | %Missed | ExtraBases | %Extra | BrokenCDS | IntactCDS | ContigN50 | ContigN90 | MaxContigLength | Blast_IntactCDS |
| Abyss_k29 | 130 | 4634010 | 108 | 322 | 30 | 118 | 115 | 61835 | 1.3327 | 40405 | 0.8719 | 54 | 4266 | 95691 | 26567 | 268182 | 4267 |
| Abyss_k31 | 133 | 4626205 | 107 | 334 | 69 | 123 | 119 | 57847 | 1.2468 | 29424 | 0.636 | 57 | 4263 | 96157 | 26096 | 222425 | 4257 |
| Abyss_k33 | 135 | 4644184 | 106 | 354 | 338 | 139 | 119 | 66355 | 1.4302 | 44937 | 0.9676 | 78 | 4242 | 89001 | 24907 | 268398 | 4263 |
| CISA_Set2 | 106 | 4635648 | 93 | 321 | 131 | 117 | 104 | 55343 | 1.1928 | 39776 | 0.858 | 64 | 4256 | 113377 | 27272 | 222663 | 4268 |
| SOAP_k29 | 1373 | 4582756 | 479 | 48 | 0 | 466 | 415 | 124372 | 2.6806 | 7247 | 0.1581 | 100 | 4220 | 17892 | 5276 | 103369 | 4146 |
| SOAP_k31 | 1295 | 4583165 | 519 | 56 | 0 | 510 | 466 | 121606 | 2.621 | 9201 | 0.2008 | 121 | 4199 | 17003 | 4286 | 77302 | 4094 |
| SOAP_k33 | 2170 | 4608265 | 1519 | 105 | 0 | 1470 | 1380 | 126273 | 2.7216 | 41165 | 0.8933 | 507 | 3813 | 5391 | 1449 | 22953 | 3379 |
| CISA_Set3 | 440 | 4532470 | 383 | 35 | 0 | 379 | 338 | 133920 | 2.8864 | 5809 | 0.1282 | 88 | 4232 | 23332 | 6264 | 103369 | 4165 |
| CISA_Set2&3 | 105 | 4636952 | 91 | 344 | 159 | 117 | 103 | 57192 | 1.2327 | 40304 | 0.8692 | 60 | 4260 | 113377 | 27272 | 222663 | 4269 |
| CISA_Set_1_2_3_2&3 | 72 | 4637760 | 72 | 554 | 53 | 112 | 100 | 43060 | 0.9281 | 37481 | 0.8082 | 44 | 4276 | 115185 | 35678 | 310691 | 4291 |
- As can be seen here, CISA can successfully integrate the sets of contigs and reduce the number of contigs. However, in comparison with integrating assemblies generated from different assemblers (Set 1), varying assemble parameters is less efficient (in the case of Set 2 or Set3) in completing the genome.
- Scaffold the contigs using SSPACE
- Since we have the paired-end reads of E. coli, it is possible to assess the order, distance and orientation of contigs and combine them into scaffolds. We, therefore, used SSPACE to scaffold the contigs and quantified the scaffolds by Mauve assembly metrics.
| Name | NumContigs | NumAssemblyBases | DCJ_Distance | NumMisCalled | NumUnCalled | NumGapsRef | NumGapsAssembly | TotalBasesMissed | %Missed | ExtraBases | %Extra | BrokenCDS | IntactCDS | ContigN50 | MaxContigLength | Blast_IntactCDS |
| CISA+SSPACE | 69 | 4627867 | 70 | 237 | 50 | 90 | 91 | 52150 | 1.124 | 37320 | 0.8064 | 44 | 4276 | 134584 | 418148 | 4290 |
| Abyss+SSPACE | 101 | 4627104 | 89 | 393 | 735 | 114 | 119 | 54956 | 1.1845 | 33747 | 0.7293 | 57 | 4263 | 107040 | 268750 | 4272 |
| minimus2+SSPACE | 64 | 4608774 | 64 | 337 | 54 | 93 | 76 | 75502 | 1.6273 | 36021 | 0.7816 | 49 | 4271 | 150458 | 420117 | 4268 |
- The results show:
- The integrated contigs output by CISA can be scaffolded by SSPACE to a limited extent (from 72 to 69), which suggests that our CISA can indeed integrate the sequence information from different assemblies.
- To introduce the paired-end reads to the pre-assembled contigs generated by Abyss using SSPACE (Abyss+SSPACE) can only reduce the number of contigs from 133 to 101, smaller than the effect made by CISA (from 133 to 72), which suggests that contig integration prior to scaffolding can further enhance the result.
- The problem of misassembled contigs generated by minimus2 is not yet solved by SSPACE, which suggests that we should integrate contigs with caution.
