
 |
| | | Home | | | Draft reconstruction | | | About | | | How-to | | | Contact | | | Supplementary | | |
MrBac
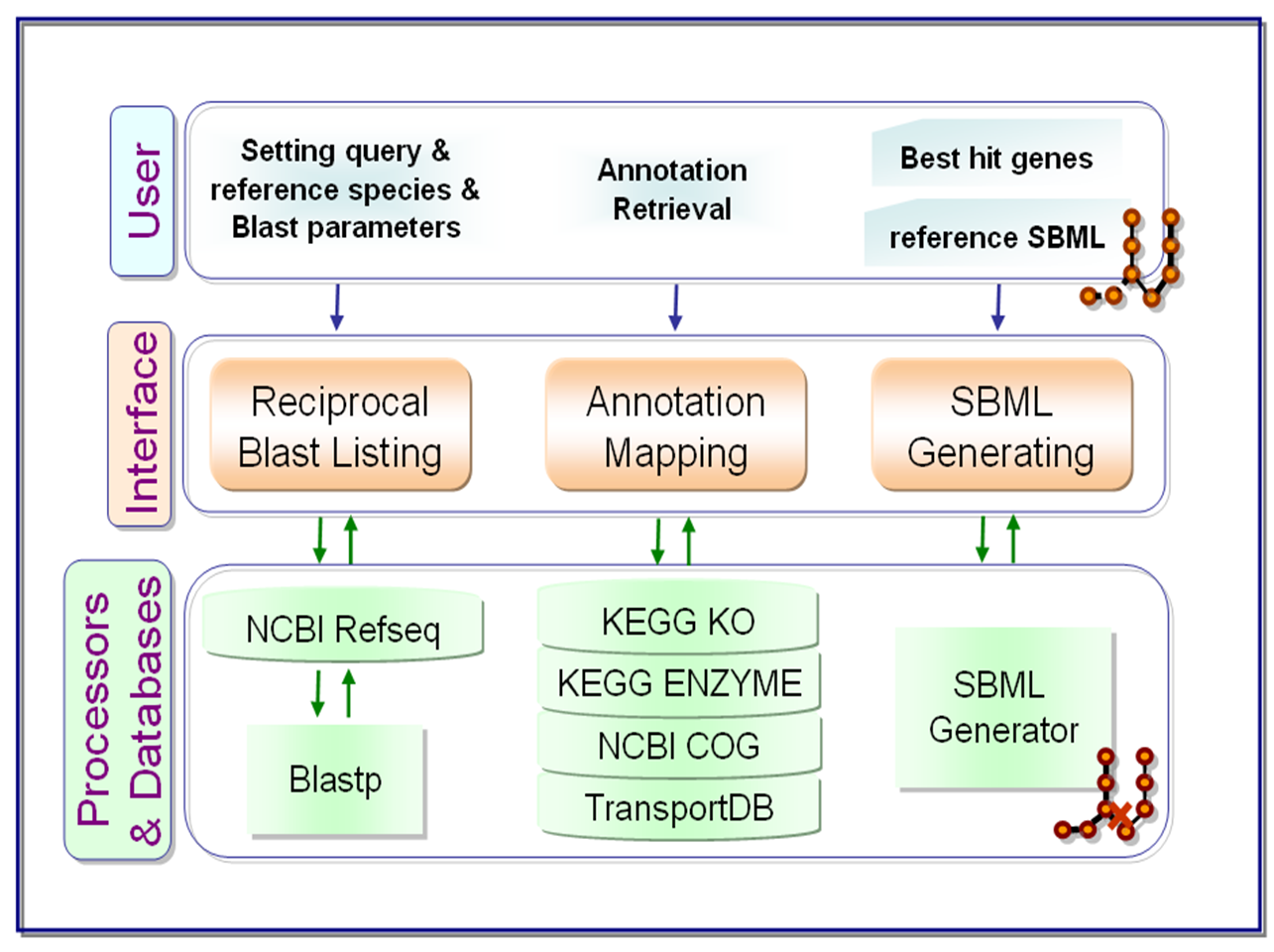
facilitates bacterial genome-scale metabolic network reconstruction through genome comparison by
(i) performing reciprocal blast on query and reference
bacterial ORFs of user’s choice,
(ii) retrieving gene annotations extracted from metabolic related databases with specified blast parameter settings and
(iii) reconstructing metabolic network of query species from
reference network distributed in Systems Biology Markup Language (SBML).
By utilizing published reconstructed metabolic networks in SBML, MrBac provides an automated draft genome-scale generation service benefiting the systems biology research community by minimizing the time spent from genome ORFs to draft genome-scale metabolic networks.
The user can select a bacterial species of interest from the menu with one reference species at a time and select a desired e-value and a percent identity as the thresholds for the reciprocal blastp best ORF hits. Further parameter adjustments can be done to obtain filtered best hits before the retrieving of metabolic gene annotations extracted from KEGG, NCBI COG and TransportDB. Draft metabolic network generation requires a list of reciprocal blast best hits as well as the reference SBML file to proceed. A draft in SBML format can be saved as the final output and can be further curated and edited in flux balance analysis softwares.
| Organism | Version | Publication | Metabolic network file | Note |
| Escherichia coli str. K-12 substr. MG1655 | iAF1260 | Feist et al. | SBML | |
| Mycoplasma genitalium | iPS189 | Suthers et al. | SBML | MG### -> MG_### |
| Thermotoga maritima MSB8 | Zhang et al. | SBML | TM_#### -> TM#### | |
| Salmonella | iMA945 | AbuOun et al. | SBML | jbc.M109.005868-7.txt -> iMA945.xml |
| Staphylococcus aureus N315 | iSB619 | Becker and Palsson. | SBML | Exported from BiGG |
| Helicobacter pylori strain 26695 | iIT341 | Thiele et al. | SBML | Exported from BiGG |
Division of Biostatistics and Bioinformatics,
Institute of Population Health Sciences,
National Health Research Institutes, Zhunan, Taiwan
All Rights Reserved
Last Updated: 04/27/2011