PBcR
The latest version of PBcR pipeline was released in Celera Assembler wgs-8.2
| Contents |
|---|
PBcR pipeline
The latest Celera Assembler integrated five steps PBcR pipeline to a single executive file "PBcR". We used different parameter setting, with and without genomeSize and pbCNS, to assemble Dataset 5 to Dataset 9
Dataset 5 (E. coli K-12 MG1655, 17 SMRT cells)
We used all SMRT cells and randomly selected four, six and eight SMRT cells three times for each, and evaluated the assemblies by QUAST against the reference genome (NC_000913) and Ec_gene_list.
PBcR -pbCNS -length 500 -partitions 200 -l ecoli -s pacbio.spec -fastq filtered_subreads.fastq PBcR -pbCNS -length 500 -partitions 200 -l ecoli -s pacbio.spec -fastq filtered_subreads.fastq genomeSize=4650000 PBcR -pbCNS -length 500 -partitions 200 -l ecoli -s pacbio.spec -fastq filtered_subreads.fastq genomeSize=4185000 PBcR -pbCNS -length 500 -partitions 200 -l ecoli -s pacbio.spec -fastq filtered_subreads.fastq genomeSize=5115000 PBcR -pbCNS -length 500 -partitions 200 -l ecoli -s pacbio.spec -fastq filtered_subreads.fastq genomeSize=3720000 PBcR -pbCNS -length 500 -partitions 200 -l ecoli -s pacbio.spec -fastq filtered_subreads.fastq genomeSize=5580000
Performance
without genomeSize(more detail)
| Statistics without reference | All Data | 4 SMRT cells : 1st Set | 4 SMRT cells : 2nd Set | 4 SMRT cells : 3rd Set | 6 SMRT cells : 1st Set | 6 SMRT cells : 2nd Set | 6 SMRT cells : 3rd Set | 8 SMRT cells : 1st Set | 8 SMRT cells : 2nd Set | 8 SMRT cells : 3rd Set |
| # contigs | 1 | 4 | 1 | 6 | 1 | 1 | 1 | 1 | 1 | 1 |
| Largest contig | 4649001 | 3622531 | 4647261 | 3760960 | 4648404 | 4654462 | 4647823 | 4649251 | 4648711 | 4648069 |
| Total length | 4649001 | 4638443 | 4647261 | 4661578 | 4648404 | 4654462 | 4647823 | 4649251 | 4648711 | 4648069 |
| N50 | 4649001 | 3622531 | 4647261 | 3760960 | 4648404 | 4654462 | 4647823 | 4649251 | 4648711 | 4648069 |
| Misassemblies | ||||||||||
| # misassemblies | 10 | 8 | 8 | 10 | 8 | 8 | 10 | 9 | 8 | 8 |
| Misassembled contigs length | 4649001 | 3622531 | 4647261 | 3864655 | 4648404 | 4654462 | 4647823 | 4649251 | 4648711 | 4648069 |
| Mismatches | ||||||||||
| # mismatches per 100kbp | 0.19 | 0.5 | 0.26 | 0.97 | 0.22 | 0.280 | 0.26 | 0.26 | 0.09 | 0.13 |
| # indels per 100kbp | 1.64 | 26.42 | 20.150 | 15.86 | 6.66 | 10.76 | 5.82 | 4.1 | 5.3 | 6.68 |
| # N's per 100kbp | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Genome Statistics | ||||||||||
| Genome fraction(%) | 100 | 99.942 | 100 | 99.999 | 100 | 100 | 100 | 100 | 100 | 100 |
| Duplication ratio | 1.002 | 1 | 1.002 | 1.006 | 1.002 | 1.003 | 1.002 | 1.002 | 1.002 | 1.002 |
| # genes | 4494 +3 part | 4488 +8 part | 4494 +3 part | 4492 +5 part | 4494 +3 part | 4495 +2 part | 4494 +3 part | 4494 +3 part | 4494 +3 part | 4494 +3 part |
| NGA50 | 3026385 | 907188 | 949093 | 880427 | 3026217 | 3026142 | 3026271 | 1257063 | 2856677 | 2856668 |
| Running Time | 6hr 14m | 30m 13s | 39m 45s | 33m 56s | 1hr 5m | 48m 4s | 1hr 2m | 1hr 22m | 1hr 31m | 1hr 26m |
genomeSize = 4650000 (more detail)
| Statistics without reference | All Data | 4 SMRT cells : 1st Set | 4 SMRT cells : 2nd Set | 4 SMRT cells : 3rd Set | 6 SMRT cells : 1st Set | 6 SMRT cells : 2nd Set | 6 SMRT cells : 3rd Set | 8 SMRT cells : 1st Set | 8 SMRT cells : 2nd Set | 8 SMRT cells : 3rd Set |
| # contigs | 1 | 4 | 1 | 6 | 1 | 1 | 1 | 1 | 1 | 1 |
| Largest contig | 4 648 864 | 3 621 580 | 4 639 780 | 2 212 593 | 4 651 575 | 4 645 728 | 4 647 827 | 4 649 239 | 4 648 687 | 4 648 099 |
| Total length | 4 648 864 | 4 602 589 | 4 639 780 | 4 661 453 | 4 651 575 | 4 645 728 | 4 647 827 | 4 649 239 | 4 648 687 | 4 648 099 |
| N50 | 4 648 864 | 3 621 580 | 4 639 780 | 887 256 | 4 651 575 | 4 645 728 | 4 647 827 | 4 649 239 | 4 648 687 | 4 648 099 |
| Misassemblies | ||||||||||
| # misassemblies | 10 | 8 | 8 | 8 | 8 | 8 | 10 | 9 | 8 | 8 |
| Misassembled contigs length | 4 648 864 | 3 621 580 | 4 639 780 | 3 857 726 | 4 651 575 | 4 645 728 | 4 647 827 | 4 649 239 | 4 648 687 | 4 648 099 |
| Mismatches | ||||||||||
| # mismatches per 100kbp | 0.19 | 0.54 | 0.37 | 0.34 | 0.17 | 0.09 | 0.15 | 0.34 | 0.09 | 0.11 |
| # indels per 100kbp | 1.59 | 24.54 | 22.42 | 15.82 | 6.38 | 10.22 | 5.63 | 4.38 | 5.32 | 6.06 |
| # N's per 100kbp | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Genome Statistics | ||||||||||
| Genome fraction(%) | 100 | 99.169 | 99.965 | 99.981 | 100 | 100 | 100 | 100 | 100 | 100 |
| Duplication ratio | 1.002 | 1 | 1 | 1.003 | 1.003 | 1.001 | 1.002 | 1.002 | 1.002 | 1.002 |
| # genes | 4494+3 part | 4458+8 part | 4491+5 part | 4491+6 part | 4494+3 part | 4494+3 part | 4494+3 part | 4494+3 part | 4494+3 part | 4493+3 part |
| NGA50 | 3 026 386 | 907 076 | 1 097 763 | 880 448 | 3 026 238 | 2 603 928 | 3 026 270 | 1 257 068 | 2 856 673 | 2 856 687 |
| Running Time | 12hr 14m | 26m 16s | 35m 50s | 33m 3s | 53m 36s | 48m 9s | 2hr 21m | 2hr 43s | 1hr 2m | 53m 3s |
genomeSize=4185000 (more detail)
| Statistics without reference | All Data | 4 SMRT cells : 1st Set | 4 SMRT cells : 2nd Set | 4 SMRT cells : 3rd Set | 6 SMRT cells : 1st Set | 6 SMRT cells : 2nd Set | 6 SMRT cells : 3rd Set | 8 SMRT cells : 1st Set | 8 SMRT cells : 2nd Set | 8 SMRT cells : 3rd Set |
| # contigs | 2 | 6 | 1 | 6 | 1 | 1 | 1 | 1 | 1 | 1 |
| Largest contig | 4 644 985 | 3 621 592 | 4 639 809 | 2 212 597 | 4 651 574 | 4 645 691 | 4 647 833 | 4 649 261 | 4 648 735 | 4 648 123 |
| Total length | 4 656 267 | 4 640 061 | 4 639 809 | 4 650 332 | 4 651 574 | 4 645 691 | 4 647 833 | 4 649 261 | 4 648 735 | 4 648 123 |
| N50 | 4 644 985 | 3 621 592 | 4 639 809 | 887 254 | 4 651 574 | 4 645 691 | 4 647 833 | 4 649 261 | 4 648 735 | 4 648 123 |
| Misassemblies | ||||||||||
| # misassemblies | 12 | 8 | 8 | 8 | 8 | 8 | 10 | 9 | 8 | 8 |
| Misassembled contigs length | 4 656 267 | 3 621 592 | 4 639 809 | 3 857 728 | 4 651 574 | 4 645 691 | 4 647 833 | 4 649 261 | 4 648 735 | 4 648 123 |
| Mismatches | ||||||||||
| # mismatches per 100kbp | 0.24 | 0.26 | 0.32 | 0.39 | 0.19 | 0.09 | 0.22 | 0.3 | 0.06 | 0.06 |
| # indels per 100kbp | 2.11 | 27.51 | 22.29 | 15.74 | 6.36 | 10.91 | 5.35 | 3.51 | 4.94 | 5.6 |
| # N's per 100kbp | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Genome Statistics | ||||||||||
| Genome fraction(%) | 100 | 99.9 | 99.965 | 99.981 | 100 | 100 | 100 | 100 | 100 | 100 |
| Duplication ratio | 1.004 | 1.001 | 1 | 1.003 | 1.003 | 1.001 | 1.002 | 1.002 | 1.002 | 1.002 |
| # genes | 4494+3 part | 4486+8 part | 4491+5 part | 4491+6 part | 4494+3 part | 4494+3 part | 4494+3 part | 4494+3 part | 4494+3 part | 4494+3 part |
| NGA50 | 3 026 366 | 904 084 | 1 097 787 | 880 446 | 3 026 242 | 2 603 887 | 3 026 279 | 1 257 060 | 2 856 681 | 2 856 711 |
| Running Time | 10hr 1m | 24m 51s | 30m 28s | 27m 44s | 43m 13s | 34m 39s | 41m 29s | 49m 7s | 52m 10s | 50m 29s |
genomeSize=5115000 (more detail)
| Statistics without reference | All Data | 4 SMRT cells : 1st Set | 4 SMRT cells : 2nd Set | 4 SMRT cells : 3rd Set | 6 SMRT cells : 1st Set | 6 SMRT cells : 2nd Set | 6 SMRT cells : 3rd Set | 8 SMRT cells : 1st Set | 8 SMRT cells : 2nd Set | 8 SMRT cells : 3rd Set |
| # contigs | 1 | 5 | 1 | 6 | 1 | 2 | 1 | 1 | 1 | 1 |
| Largest contig | 4 649 007 | 3 621 673 | 4 639 792 | 2 212 738 | 4 648 429 | 4 645 722 | 4 647 825 | 4 649 266 | 4 648 723 | 4 648 107 |
| Total length | 4 649 007 | 4 636 546 | 4 639 792 | 4 650 567 | 4 648 429 | 4 645 722 | 4 647 825 | 4 649 266 | 4 648 723 | 4 648 107 |
| N50 | 4 649 007 | 3 621 673 | 4 639 792 | 887 256 | 4 648 429 | 4 645 722 | 4 647 825 | 4 649 266 | 4 648 723 | 4 648 107 |
| Misassemblies | ||||||||||
| # misassemblies | 10 | 8 | 8 | 8 | 8 | 8 | 10 | 9 | 8 | 8 |
| Misassembled contigs length | 4 649 007 | 3 621 673 | 4 639 792 | 3 857 870 | 4 648 429 | 4 645 722 | 4 647 825 | 4 649 266 | 4 648 723 | 4 648 107 |
| Mismatches | ||||||||||
| # mismatches per 100kbp | 0.17 | 0.32 | 0.41 | 0.37 | 0.19 | 0.11 | 0.15 | 0.39 | 0.06 | 0.11 |
| # indels per 100kbp | 1.53 | 25.72 | 22.34 | 15.84 | 6.21 | 10.32 | 5.63 | 3.53 | 5.17 | 6.06 |
| # N's per 100kbp | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Genome Statistics | ||||||||||
| Genome fraction(%) | 100 | 99.9 | 99.965 | 99.981 | 100 | 100 | 100 | 100 | 100 | 100 |
| Duplication ratio | 1.002 | 1 | 1 | 1.003 | 1.002 | 1.001 | 1.002 | 1.002 | 1.002 | 1.002 |
| # genes | 4494 +3 part | 4486 +8 part | 4491 +5 part | 4491 +6 part | 4494 +3 part | 4494 +3 part | 4494 +3 part | 4494 +3 part | 4494 +3 part | 4494 +3 part |
| NGA50 | 3 026 390 | 907 197 | 1 097 774 | 880 448 | 3 026 239 | 2 603 918 | 3 026 271 | 1 257 073 | 2 856 674 | 2 856 693 |
| Running Time | ||||||||||
| Running Time | 2hr 24m | 26m 41s | 33m 14s | 30m 4s | 47m 8s | 37m 26s | 46m 5s | 54m 46s | 59m 16s | 56m 19s |
genomeSize=3720000 (more detail)
| Statistics without reference | All Data | 4 SMRT cells : 1st Set | 4 SMRT cells : 2nd Set | 4 SMRT cells : 3rd Set | 6 SMRT cells : 1st Set | 6 SMRT cells : 2nd Set | 6 SMRT cells : 3rd Set | 8 SMRT cells : 1st Set | 8 SMRT cells : 2nd Set | 8 SMRT cells : 3rd Set |
| # contigs | 1 | 6 | 1 | 6 | 1 | 1 | 1 | 1 | 1 | 1 |
| Largest contig | 4 648 264 | 3 621 566 | 4 639 815 | 2 212 780 | 4 651 567 | 4 654 368 | 4 647 818 | 4 649 190 | 4 648 720 | 4 648 103 |
| Total length | 4 648 264 | 4 640 025 | 4 639 815 | 4 649 418 | 4 651 567 | 4 654 368 | 4 647 818 | 4 649 190 | 4 648 720 | 4 648 103 |
| N50 | 4 648 264 | 3 621 566 | 4 639 815 | 887 256 | 4 651 567 | 4 654 368 | 4 647 818 | 4 649 190 | 4 648 720 | 4 648 103 |
| Misassemblies | ||||||||||
| # misassemblies | 10 | 8 | 8 | 8 | 8 | 8 | 10 | 9 | 8 | 8 |
| Misassembled contigs length | 4 648 264 | 3 621 566 | 4 639 815 | 3 857 911 | 4 651 567 | 4 654 368 | 4 647 818 | 4 649 190 | 4 648 720 | 4 648 103 |
| Mismatches | ||||||||||
| # mismatches per 100kbp | 0.17 | 0.28 | 0.39 | 0.43 | 0.22 | 0.28 | 0.19 | 0.37 | 0.13 | 0.11 |
| # indels per 100kbp | 1.72 | 25.24 | 22.1 | 15.82 | 6.79 | 11.7 | 5.43 | 4.440 | 5.24 | 6.19 |
| # N's per 100kbp | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Genome Statistics | ||||||||||
| Genome fraction(%) | 100 | 99.9 | 99.965 | 99.981 | 100 | 100 | 100 | 100 | 100 | 100 |
| Duplication ratio | 1.002 | 1.001 | 1 | 1.002 | 1.003 | 1.003 | 1.002 | 1.002 | 1.002 | 1.002 |
| # genes | 4494+3 part | 4486+8 part | 4491+5 part | 4490+7 part | 4494+3 part | 4495+2 part | 4494+3 part | 4494+3 part | 4494+3 part | 4494+3 part |
| NGA50 | 3 026 382 | 907 054 | 1 097 788 | 880 448 | 3 026 242 | 3 026 108 | 3 026 274 | 1 257 032 | 2 856 687 | 2 856 706 |
| Running Time | 1hr 45m | 24m 2s | 28m 45s | 26m 23s | 40m 12s | 32m 31s | 38m 38s | 45m 55s | 48m 40s | 46m 47s |
genomeSize=5580000 (more detail)
| Statistics without reference | All Data | 4 SMRT cells : 1st Set | 4 SMRT cells : 2nd Set | 4 SMRT cells : 3rd Set | 6 SMRT cells : 1st Set | 6 SMRT cells : 2nd Set | 6 SMRT cells : 3rd Set | 8 SMRT cells : 1st Set | 8 SMRT cells : 2nd Set | 8 SMRT cells : 3rd Set |
| # contigs | 1 | 4 | 1 | 6 | 1 | 1 | 1 | 1 | 1 | 1 |
| Largest contig | 4 649 007 | 3 622 415 | 4 639 778 | 3 100 853 | 4 651 559 | 4 652 494 | 4 647 813 | 4 649 253 | 4 648 694 | 4 648 107 |
| Total length | 4 649 007 | 4 638 349 | 4 639 778 | 4 660 255 | 4 651 559 | 4 652 494 | 4 647 813 | 4 649 253 | 4 648 694 | 4 648 107 |
| N50 | 4 649 007 | 3 622 415 | 4 639 778 | 3 100 853 | 4 651 559 | 4 652 494 | 4 647 813 | 4 649 253 | 4 648 694 | 4 648 107 |
| Misassemblies | ||||||||||
| # misassemblies | 10 | 8 | 8 | 8 | 8 | 9 | 10 | 9 | 8 | 8 |
| Misassembled contigs length | 4 649 007 | 3 622 415 | 4 639 778 | 3 858 736 | 4 651 559 | 4 652 494 | 4 647 813 | 4 649 253 | 4 648 694 | 4 648 107 |
| Mismatches | ||||||||||
| # mismatches per 100kbp | 0.19 | 0.43 | 0.37 | 0.34 | 0.15 | 0.09 | 0.15 | 0.37 | 0.11 | 0.11 |
| # indels per 100kbp | 1.51 | 25.84 | 22.42 | 15.88 | 6.4 | 10.35 | 5.73 | 3.66 | 5.22 | 6.06 |
| # N's per 100kbp | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Genome Statistics | ||||||||||
| Genome fraction(%) | 100 | 99.937 | 99.965 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| Duplication ratio | 1.002 | 1 | 1 | 1.004 | 1.003 | 1.003 | 1.002 | 1.002 | 1.002 | 1.002 |
| # genes | 4494+3 part | 4488+8 part | 4491+5 part | 4492+5 part | 4494+3 part | 4494+3 part | 4494+3 part | 4494+3 part | 4494+3 part | 4494+3 part |
| NGA50 | 3 026 391 | 907 173 | 1 097 770 | 880 460 | 3 026 223 | 1 252 894 | 3 026 257 | 1 257 068 | 2 856 688 | 2 856 692 |
| Running Time | 2hr 9m | 28m 59s | 33m 32s | 32m 21s | 50m 17s | 38m 36s | 49m 13s | 57m 6s | 1hr 24s | 59m 7s |
Dataset 6 (E.coli K-12 MG1655, 8 SMRT cells)
We used all SMRT cells and randomly selected four and six SMRT cells three times for each, and evaluated the assemblies by QUAST against the reference genome (NC_000913) and Ec_gene_list.
PBcR -pbCNS -length 500 -partitions 200 -l ecoli -s pacbio.spec -fastq filtered_subreads.fastq genomeSize=4650000 PBcR -pbCNS -length 500 -partitions 200 -l ecoli -s pacbio.spec -fastq filtered_subreads.fastq genomeSize=4185000 PBcR -pbCNS -length 500 -partitions 200 -l ecoli -s pacbio.spec -fastq filtered_subreads.fastq genomeSize=5115000 PBcR -pbCNS -length 500 -partitions 200 -l ecoli -s pacbio.spec -fastq filtered_subreads.fastq genomeSize=3720000 PBcR -pbCNS -length 500 -partitions 200 -l ecoli -s pacbio.spec -fastq filtered_subreads.fastq genomeSize=5580000
Performance
genomeSize=4650000 (more detail)
| Statistics without reference | All Data | 4 SMRT cells : 1st Set | 4 SMRT cells : 2nd Set | 4 SMRT cells : 3rd Set | 6 SMRT cells : 1st Set | 6 SMRT cells : 2nd Set | 6 SMRT cells : 3rd Set |
| # contigs | 2 | 51 | 44 | 76 | 6 | 6 | 6 |
| Largest contig | 3 835 938 | 300 354 | 500 180 | 250 542 | 2 045 145 | 2 044 223 | 2 542 485 |
| Total length | 4 640 874 | 4 437 792 | 4 476 210 | 4 297 112 | 4 636 889 | 4 635 531 | 4 645 642 |
| N50 | 3 835 938 | 105 841 | 117 447 | 69 771 | 1 293 614 | 1 522 526 | 2 542 485 |
| Misassemblies | |||||||
| # misassemblies | 8 | 6 | 7 | 6 | 7 | 8 | 9 |
| Misassembled contigs length | 3 835 938 | 678 934 | 950 771 | 435 912 | 3 567 800 | 3 623 564 | 3 630 045 |
| Mismatches | |||||||
| # mismatches per 100kbp | 0.19 | 1.65 | 1.61 | 3.74 | 0.35 | 0.24 | 0.35 |
| # indels per 100kbp | 4.81 | 56.36 | 39.36 | 65.58 | 9.72 | 9.72 | 9.84 |
| # N's per 100kbp | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Genome Statistics | |||||||
| Genome fraction(%) | 99.938 | 95.61 | 96.483 | 92.288 | 99.828 | 99.762 | 99.86 |
| Duplication ratio | 1.001 | 1.005 | 1.006 | 1.007 | 1.001 | 1.001 | 1.003 |
| # genes | 4490 +5 part | 4229 +71 part | 4288 +65 part | 4094 +105 part | 4481 +10 part | 4475 +11 part | 4485 +8 part |
| NGA50 | 949 276 | 89 654 | 111 935 | 53 222 | 857 569 | 857 671 | 859 217 |
| Running Time | 35m 5s | 17m 43s | 18m 14s | 16m 12s | 26m 17s | 26m 9s | 27m 2s |
genomeSize=4185000 (more detail)
| Statistics without reference | All Data | 4 SMRT cells : 1st Set | 4 SMRT cells : 2nd Set | 4 SMRT cells : 3rd Set | 6 SMRT cells : 1st Set | 6 SMRT cells : 2nd Set | 6 SMRT cells : 3rd Set |
| # contigs | 2 | 50 | 45 | 75 | 7 | 6 | 7 |
| Largest contig | 3 835 933 | 300 354 | 500 170 | 250 539 | 2 044 974 | 2 044 226 | 2 177 146 |
| Total length | 4 640 868 | 4 437 828 | 4 476 213 | 4 279 289 | 4 636 194 | 4 635 257 | 4 641 795 |
| N50 | 3 835 933 | 108 835 | 113 085 | 69 768 | 805 910 | 1 522 465 | 1 026 972 |
| Misassemblies | |||||||
| # misassemblies | 8 | 6 | 7 | 6 | 8 | 8 | 8 |
| Misassembled contigs length | 3 835 933 | 678 928 | 950 762 | 435 912 | 3 005 683 | 3 623 505 | 3 204 118 |
| Mismatches | |||||||
| # mismatches per 100kbp | 0.19 | 1.69 | 1.61 | 3.92 | 0.32 | 0.24 | 0.26 |
| # indels per 100kbp | 5.39 | 55.34 | 39.72 | 66.040 | 10.84 | 10.07 | 10.1 |
| # N's per 100kbp | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Genome Statistics | |||||||
| Genome fraction(%) | 99.938 | 95.612 | 96.481 | 91.936 | 99.78 | 99.757 | 99.832 |
| Duplication ratio | 1.001 | 1 | 1 | 1.005 | 1.002 | 1.002 | 1.002 |
| # genes | 4490 +5 part | 4230 +71 part | 4287 +65 part | 4081 +106 part | 4476 +11 part | 4475 +11 part | 4484 +9 part |
| NGA50 | 949 260 | 91 700 | 98 453 | 53 221 | 805 910 | 857 615 | 770 513 |
| Running Time | 33m 51s | 17m 26s | 18m 7s | 16m 23s | 25m 54s | 24m 48s | 26m 8s |
genomeSize = 5115000 (more detail)
| Statistics without reference | All Data | 4 SMRT cells : 1st Set | 4 SMRT cells : 2nd Set | 4 SMRT cells : 3rd Set | 6 SMRT cells : 1st Set | 6 SMRT cells : 2nd Set | 6 SMRT cells : 3rd Set |
| # contigs | 2 | 51 | 44 | 78 | 6 | 6 | 6 |
| Largest contig | 3 835 925 | 300 350 | 500 182 | 250 540 | 2 045 293 | 2 044 230 | 2 542 455 |
| Total length | 4 640 858 | 4 437 775 | 4 476 309 | 4 283 745 | 4 637 036 | 4 631 106 | 4 645 623 |
| N50 | 3 835 925 | 105 842 | 117 459 | 60 082 | 1 293 612 | 1 522 478 | 2 542 455 |
| Misassemblies | |||||||
| # misassemblies | 8 | 6 | 7 | 6 | 7 | 7 | 9 |
| Misassembled contigs length | 3 835 925 | 678 925 | 950 766 | 435 899 | 356 7945 | 3 566 708 | 3 630 015 |
| Mismatches | |||||||
| # mismatches per 100kbp | 0.19 | 1.62 | 1.56 | 3.7 | 0.35 | 0.24 | 0.3 |
| # indels per 100kbp | 4.940 | 56.09 | 39.38 | 66.69 | 10.75 | 10.09 | 10.66 |
| # N's per 100kbp | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Genome Statistics | |||||||
| Genome fraction(%) | 99.938 | 95.61 | 96.485 | 91.98 | 99.831 | 99.757 | 99.86 |
| Duplication ratio | 1.001 | 1 | 1 | 1.005 | 1.001 | 1.001 | 1.003 |
| # genes | 4490 +5 part | 4229 +71 part | 4288 +64 part | 4085 +107 part | 4481 +10 part | 4474 +12 part | 4485 +8 part |
| NGA50 | 949 271 | 89 653 | 111 937 | 53 221 | 857 567 | 857 628 | 859 219 |
| Running Time | 36m 41s | 17m 52s | 18m 22s | 16m 44s | 27m 49s | 26m 54s | 27m 59s |
genomeSize=3720000 (more detail)
| Statistics without reference | All Data | 4 SMRT cells : 1st Set | 4 SMRT cells : 2nd Set | 4 SMRT cells : 3rd Set | 6 SMRT cells : 1st Set | 6 SMRT cells : 2nd Set | 6 SMRT cells : 3rd Set |
| # contigs | 2 | 51 | 45 | 74 | 10 | 7 | 7 |
| Largest contig | 3 835 918 | 300 357 | 499 793 | 250 543 | 1 068 976 | 2 044 231 | 2 177 132 |
| Total length | 4 640 911 | 4 429 199 | 4 462 499 | 4 252 687 | 4 637 980 | 4 633 643 | 4 641 751 |
| N50 | 3 835 918 | 108 839 | 113 115 | 69 767 | 674 174 | 1 522 440 | 1 026 958 |
| Misassemblies | |||||||
| # misassemblies | 8 | 6 | 7 | 5 | 8 | 8 | 8 |
| Misassembled contigs length | 3 835 918 | 677 539 | 950 434 | 397 273 | 3 005 363 | 3 623 486 | 3 204 090 |
| Mismatches | |||||||
| # mismatches per 100kbp | 0.24 | 1.67 | 1.57 | 3.3 | 0.78 | 0.3 | 0.3 |
| # indels per 100kbp | 5.97 | 55.08 | 39.44 | 66 | 11.98 | 11.35 | 10.28 |
| # N's per 100kbp | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Genome Statistics | |||||||
| Genome fraction(%) | 99.938 | 95.44 | 96.187 | 91.369 | 99.687 | 99.723 | 99.832 |
| Duplication ratio | 1.001 | 1 | 1 | 1.004 | 1.004 | 1.002 | 1.002 |
| # genes | 4490 +5 part | 4221 +75 part | 4274 +65 part | 4056 +105 part | 4467 +13 part | 4471 +13 part | 4484 +9 part |
| NGA50 | 949 257 | 91 700 | 98 451 | 53 254 | 618 553 | 857 595 | 770 508 |
| Running Time | 33m 1s | 17m 3s | 17m 19s | 16m 9s | 24m 53s | 24m 12s | 25m 13s |
genomeSize=5580000 (more detail)
| Statistics without reference | All Data | 4 SMRT cells : 1st Set | 4 SMRT cells : 2nd Set | 4 SMRT cells : 3rd Set | 6 SMRT cells : 1st Set | 6 SMRT cells : 2nd Set | 6 SMRT cells : 3rd Set |
| # contigs | 4 | 52 | 44 | 76 | 6 | 6 | 3 |
| Largest contig | 3 161 528 | 300 351 | 500 181 | 250 537 | 2 045 299 | 2 044 231 | 3 622 857 |
| Total length | 4 662 182 | 4 441 307 | 4 483 671 | 4 298 734 | 4 637 078 | 4 631 147 | 4 635 596 |
| N50 | 3 161 528 | 93 960 | 113 089 | 63 438 | 1 293 638 | 1 522 515 | 3 622 857 |
| Misassemblies | |||||||
| # misassemblies | 8 | 6 | 7 | 7 | 7 7 | 8 | |
| Misassembled contigs length | 3 161 528 | 678 927 | 950 772 | 526 030 | 3 567 978 | 3 566 746 | 3 622 857 |
| Mismatches | |||||||
| # mismatches per 100kbp | 0.32 | 1.56 | 1.63 | 3.36 | 0.37 | 0.19 | 0.35 |
| # indels per 100kbp | 4.27 | 55.84 | 39.54 | 66.83 | 10.58 | 9.68 | 10.92 |
| # N's per 100kbp | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Genome Statistics | |||||||
| Genome fraction(%) | 99.938 | 95.614 | 96.642 | 92.463 | 99.831 | 99.757 | 99.862 |
| Duplication ratio | 1.005 | 1.001 | 1 | 1.002 | 1.001 | 1.001 | 1.001 |
| # genes | 4490 +5 part | 4230 +70 part | 4293 +65 part | 4105 +105 part | 4481 +10 part | 4474 +12 part | 4487 +6 part |
| NGA50 | 702 757 | 89 630 | 111 935 | 53 224 | 857 595 | 857 662 | 910 332 |
| Running Time | 37m 36s | 18m 7s | 18m 27s | 16m 51s | 28m 57s | 27m 36s | 28m 25s |
higher coverage bias
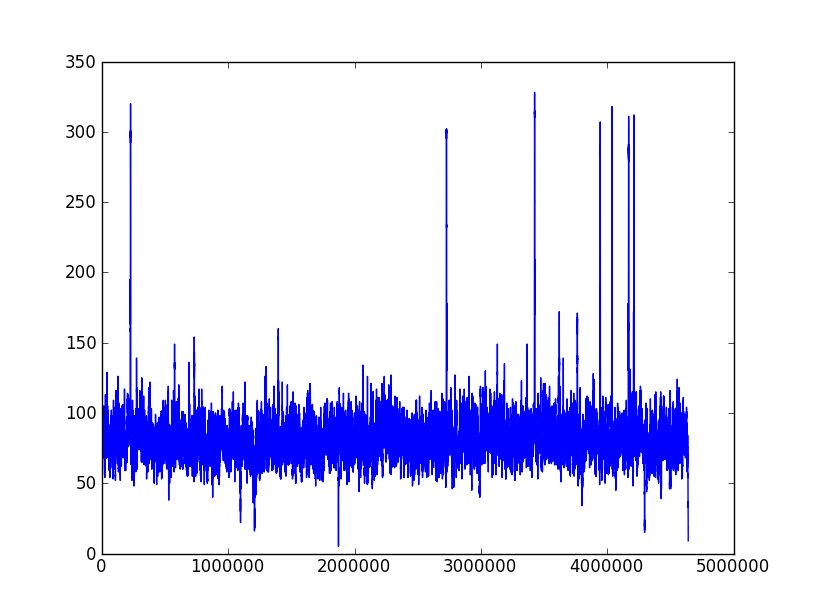
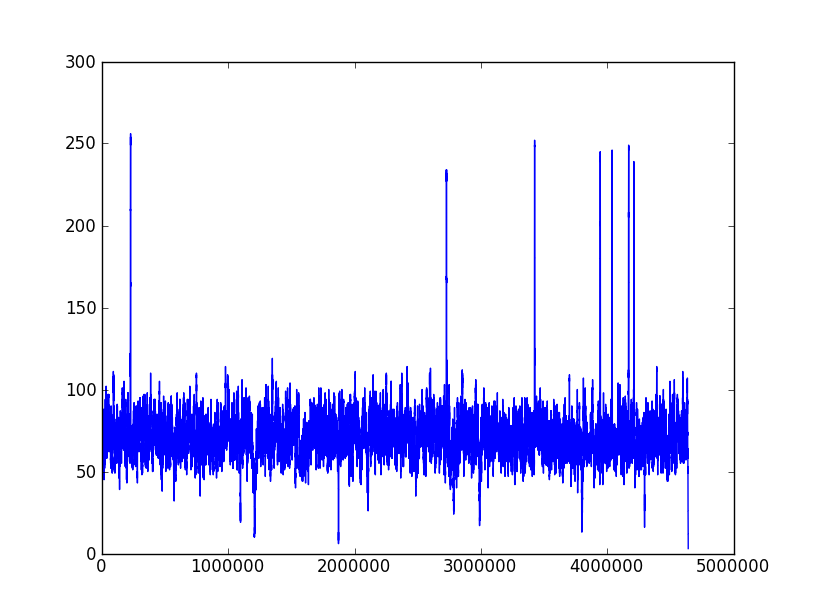
The following two pictures are the coverage distribution from eight SMRT cells of DataSet 5 and DataSet 6,and the x-axis denotes the reference genome length and the y-axis represents the coverage in each nucleotide of reference genome. These two datasets have the similar size of long reads and over 75X depth of coverage, but the dataset 6 couldn't complete genome as correctly as dataset 5. We found that there were more regions with low coverage in dataset 6 than dataset 5. The more low-coverage regions may induce the more reads couldn't be self-corrected so that there were not enough correctly overlapped information to assemble the contigs. Nevertheless, the upgraded RS II system increased the average read length to 5 Kbp (in Dataset 9) and expectedly provided average read lengths in excess of 10 Kbp with new chemistry (P6-C4). Besides, the continuously increased throughput would overcome the coverage bias.
Coverage distribution of DataSet 5
| Filtered_eight.fastq |
| seqs amount:270469 |
| seq avg len:2285.672846 |
| total:618.20 Mb |
| depth: 132.95X |
Coverage distribution of Dataset 6
| Filtered_four.fastq |
| seqs amount:187921 |
| seq avg len:3190.512705 |
| total:599.56 Mb |
| depth: 128.94X |
Dataset 7, (M. ruber DSM1279, 4 SMRT cells)
We used all SMRT cells to do assembly and evaluated the assemblies by QUAST against the reference genome (NC_013946) and Mr_gene_list.
PBcR -pbCNS -length 500 -partitions 200 -l mruber -s pacbio.spec -fastq filtered_subreads.fastq PBcR -pbCNS -length 500 -partitions 200 -l mruber -s pacbio.spec -fastq filtered_subreads.fastq genomeSize=3100000 PBcR -pbCNS -length 500 -partitions 200 -l mruber -s pacbio.spec -fastq filtered_subreads.fastq genomeSize=2790000 PBcR -pbCNS -length 500 -partitions 200 -l mruber -s pacbio.spec -fastq filtered_subreads.fastq genomeSize=3410000 PBcR -pbCNS -length 500 -partitions 200 -l mruber -s pacbio.spec -fastq filtered_subreads.fastq genomeSize=2480000 PBcR -pbCNS -length 500 -partitions 200 -l mruber -s pacbio.spec -fastq filtered_subreads.fastq genomeSize=3720000
Performance
without genomeSize (more detail)
| Statistics without reference | All Data | 3 SMRT cells : 1st Set | 3 SMRT cells : 2nd Set | 3 SMRT cells : 3rd Set | 3 SMRT cells : 4th Set |
| # contigs | 1 | 1 | 1 | 1 | 1 |
| Largest contig | 3 100 140 | 3 099 663 | 3 099 663 | 3 098 784 | 3 099 602 |
| Total length | 3 100 140 | 3 099 663 | 3 099 663 | 3 098 784 | 3 099 602 |
| N50 | 3 100 140 | 3 099 663 | 3 099 663 | 3 098 784 | 3 099 602 |
| Misassemblies | |||||
| # misassemblies | 1 | 0 | 0 | 0 | 0 |
| Misassembled contigs length | 3 100 140 | 0 | 0 | 0 | 0 |
| Mismatches | |||||
| # mismatches per 100kbp | 0.03 | 0.06 | 0.06 | 0.03 | 0.03 |
| # indels per 100kbp | 13.85 | 20.47 | 20.47 | 19.95 | 20.7 |
| # N's per 100kbp | 0 | 0 | 0 | 0 | 0 |
| Genome Statistics | |||||
| Genome fraction(%) | 99.986 | 99.986 | 99.986 | 99.986 | 99.986 |
| Duplication ratio | 1.001 | 1.001 | 1.001 | 1.001 | 1.001 |
| # genes | 3103 + 2 part | 3103 +2 part | 3103 +2 part | 3103 +2 part | 3103 +2 part |
| NGA50 | 1 707 540 | 3 099 663 | 3 099 663 | 3 098 784 | 3 099 602 |
| Running Time | 34m 24s | 42m 32s | 37m 55s | 39m 37s | 43m 28s |
with genomeSize (more detail)
| Statistics without reference | genomeSize=3100000 | genomeSize=2790000 | genomeSize=3410000 | genomeSize=2480000 | genomeSize=3720000 |
| # contigs | 1 | 1 | 1 | 1 | 1 |
| Largest contig | 3100062 | 3100061 | 3100039 | 3100030 | 3100027 |
| Total length | 3100062 | 3100061 | 3100039 | 3100030 | 3100027 |
| N50 | 3100062 | 3100061 | 3100039 | 3100030 | 3100027 |
| Misassemblies | |||||
| # misassemblies | 0 | 0 | 0 | 0 | 0 |
| Misassembled contigs length | 0 | 0 | 0 | 0 | 0 |
| Mismatches | |||||
| # mismatches per 100kbp | 0.03 | 0.03 | 0.03 | 0.03 | 0.13 |
| # indels per 100kbp | 13.53 | 13.43 | 13.46 | 13.92 | 13.82 |
| # N's per 100kbp | 0 | 0 | 0 | 0 | 0 |
| Genome Statistics | |||||
| Genome fraction(%) | 99.986 | 99.986 | 99.986 | 99.986 | 99.986 |
| Duplication ratio | 1.001 | 1.001 | 1.001 | 1.001 | 1.001 |
| # genes | 3103 + 2part | 3103 +2 part | 3103 +2 part | 3103 +2 part | 3103 +2 part |
| NGA50 | 3100062 | 3100061 | 3100039 | 3100030 | 3100027 |
| Running Time | 34m 23s | 34m 5s | 41m | 34m 12s | 43m 56s |
Dataset 8 (P. heparinus DSM1279, 7 SMRT cells)
We used all SMRT cells and randomly selected four SMRT cells three times for each, and evaluated the assemblies by QUAST against the reference genome (NC_013061) and Ph_gene_list.
PBcR -pbCNS -length 500 -partitions 200 -l phep -s pacbio.spec -fastq filtered_subreads.fastq PBcR -pbCNS -length 500 -partitions 200 -l phep -s pacbio.spec -fastq filtered_subreads.fastq genomeSize=5170000 PBcR -pbCNS -length 500 -partitions 200 -l phep -s pacbio.spec -fastq filtered_subreads.fastq genomeSize=4653000 PBcR -pbCNS -length 500 -partitions 200 -l phep -s pacbio.spec -fastq filtered_subreads.fastq genomeSize=5687000 PBcR -pbCNS -length 500 -partitions 200 -l phep -s pacbio.spec -fastq filtered_subreads.fastq genomeSize=4136000 PBcR -pbCNS -length 500 -partitions 200 -l phep -s pacbio.spec -fastq filtered_subreads.fastq genomeSize=6204000
Performance
without genomeSize (more detail)
| Statistics without reference | All Data | 6 SMRT cells : 1st Set | 6 SMRT cells : 2nd Set | 6 SMRT cells : 3rd Set |
| # contigs | 1 | 1 | 1 | 1 |
| Largest contig | 5163845 | 5163749 | 5163778 | 5163424 |
| Total length | 5163845 | 5163749 | 5163778 | 5163424 |
| N50 | 5163845 | 5163749 | 5163778 | 5163424 |
| Misassemblies | ||||
| # misassemblies | 1 | 1 | 1 | 0 |
| Misassembled contigs length | 5163845 | 5163749 | 5163778 | 0 |
| Mismatches | ||||
| # mismatches per 100kbp | 5.85 | 5.21 | 3.39 | 3.54 |
| # indels per 100kbp | 0.64 | 1.140 | 1.18 | 2.29 |
| # N's per 100kbp | 0 | 0 | 0 | 0 |
| Genome Statistics | ||||
| Genome fraction(%) | 99.945 | 99.945 | 99.945 | 99.913 |
| Duplication ratio | 1.001 | 1.001 | 1.001 | 1 |
| # genes | 4336 + 2 part | 4336 + 2 part | 4336 + 2 part | 4335 + 3 part |
| NGA50 | 2926366 | 2926293 | 2926326 | 5163424 |
| Running Time | 1hr 11m | 53m 55s | 54m 57s | 57m 8s |
genomeSize= 5170000 bp (more detail)
| Statistics without reference | All Data | 4 SMRT cells : 1st Set | 4 SMRT cells : 2nd Set | 4 SMRT cells : 3rd Set |
| # contigs | 1 | 13 | 10 | 12 |
| Largest contig | 5163845 | 2233572 | 2243053 | 2214015 |
| Total length | 5163845 | 5144475 | 5169489 | 5153037 |
| N50 | 5163845 | 1382071 | 1271605 | 13924300 |
| Misassemblies | ||||
| # misassemblies | 1 | 0 | 1 | 0 |
| Misassembled contigs length | 5163845 | 0 | 2243053 | 0 |
| Mismatches | ||||
| # mismatches per 100kbp | 3.89 | 8.4 | 6.87 | 8.2 |
| # indels per 100kbp | 0.64 | 7.23 | 5.59 | 5.44 |
| # N's per 100kbp | 0 | 0 | 0 | 0 |
| Genome Statistics | ||||
| Genome fraction(%) | 99.945 | 99.548 | 99.977 | 99.643 |
| Duplication ratio | 1.001 | 1 | 1.002 | 1.001 |
| # genes | 4336 + 2 part | 4309 + 18 part | 4329 + 10 part | 4312 + 16 part |
| NGA50 | 2926365 | 1382071 | 1271604 | 1392430 |
| Running Time | 58m | 33m 10s | 33m 51s | 33m 36s |
genomeSize= 4653000 bp (more detail)
| Statistics without reference | All Data | 4 SMRT cells : 1st Set | 4 SMRT cells : 2nd Set | 4 SMRT cells : 3rd Set |
| # contigs | 1 | 31 | 30 | 20 |
| Largest contig | 5163845 | 1821377 | 2215586 | 2212632 |
| Total length | 5163845 | 5024237 | 5108915 | 5089645 |
| N50 | 5163845 | 360474 | 1272328 | 720504 |
| Misassemblies | ||||
| # misassemblies | 1 | 0 | 0 | 0 |
| Misassembled contigs length | 5163845 | 0 | 0 | 0 |
| Mismatches | ||||
| # mismatches per 100kbp | 3.89 | 7.39 | 7.36 | 6.56 |
| # indels per 100kbp | 0.68 | 12.09 | 10.82 | 9.33 |
| # N's per 100kbp | 0 | 0 | 0 | 0 |
| Genome Statistics | ||||
| Genome fraction(%) | 99.945 | 97.19 | 98.871 | 98.476 |
| Duplication ratio | 1.001 | 1 | 1 | 1 |
| # genes | 4336 + 2 part | 4182 + 45 part | 4259 + 40 part | 4252 + 28 part |
| NGA50 | 2926367 | 360474 | 1272327 | 720504 |
| Running Time | 55m 29s | 31m 47s | 32m 44s | 31m 4s |
genomeSize= 5687000 bp (more detail)
| Statistics without reference | All Data | 4 SMRT cells : 1st Set | 4 SMRT cells : 2nd Set | 4 SMRT cells : 3rd Set |
| # contigs | 1 | 13 | 10 | 12 |
| Largest contig | 5163844 | 2233573 | 2243055 | 2214008 |
| Total length | 5163844 | 5145570 | 5169519 | 5159845 |
| N50 | 5163844 | 1382064 | 1271645 | 1392431 |
| Misassemblies | ||||
| # misassemblies | 1 | 0 | 1 | 0 |
| Misassembled contigs length | 5163844 | 0 | 2243055 | 0 |
| Mismatches | ||||
| # mismatches per 100kbp | 2.94 | 7.99 | 6.89 | 8.17 |
| # indels per 100kbp | 0.560 | 7.09 | 5.44 | 5.49 |
| # N's per 100kbp | 0 | 0 | 0 | 0 |
| Genome Statistics | ||||
| Genome fraction(%) | 99.945 | 99.569 | 99.977 | 99.775 |
| Duplication ratio | 1.001 | 1 | 1.002 | 1.001 |
| # genes | 4336 +2 part | 4309 +18 part | 4329 +10 part | 4320 +15 part |
| NGA50 | 2926365 | 1382064 | 1271643 | 1392431 |
| Running Time | 1hr 1m | 42m 37s | 44m 46s | 42m 22s |
genomeSize = 4136000 bp (more detail)
| Statistics without reference | All Data | 4 SMRT cells : 1st Set | 4 SMRT cells : 2nd Set | 4 SMRT cells : 3rd Set |
| # contigs | 1 | 31 | 30 | 19 |
| Largest contig | 5163822 | 1821375 | 2215562 | 2212634 |
| Total length | 5163822 | 5024185 | 5108963 | 5090420 |
| N50 | 5163822 | 360474 | 1272327 | 720504 |
| Misassemblies | ||||
| # misassemblies | 1 | 0 | 0 | 2 |
| Misassembled contigs length | 5163822 | 0 | 0 | 414201 |
| Mismatches | ||||
| # mismatches per 100kbp | 3.7 | 7.37 | 7.38 | 6.8 |
| # indels per 100kbp | 0.91 | 12.23 | 10.77 | 9.39 |
| # N's per 100kbp | 0 | 0 | 0 | 0 |
| Genome Statistics | ||||
| Genome fraction(%) | 99.945 | 97.189 | 98.872 | 98.478 |
| Duplication ratio | 1.001 | 1 | 1 | 1.001 |
| # genes | 4336 +2 part | 4181 +46 part | 4260 +39 part | 4252 +28 part |
| NGA50 | 2926351 | 360474 | 1272326 | 720504 |
| Running Time | 1hr 4m | 35m 10s | 36m 54s | 35m 18s |
genomeSize= 6204000 bp (more detail)
| Statistics without reference | All Data | 4 SMRT cells : 1st Set | 4 SMRT cells : 2nd Set | 4 SMRT cells : 3rd Set |
| # contigs | 2 | 14 | 9 | 12 |
| Largest contig | 5163833 | 2233593 | 2243042 | 2214012 |
| Total length | 5163833 | 5156868 | 5169779 | 5159887 |
| N50 | 5163833 | 1382073 | 1271789 | 13924340 |
| Misassemblies | ||||
| # misassemblies | 1 | 0 | 1 | 0 |
| Misassembled contigs length | 5163833 | 0 | 2243042 | 0 |
| Mismatches | ||||
| # mismatches per 100kbp | 3.97 | 8.280 | 6.93 | 8.15 |
| # indels per 100kbp | 0.64 | 7.33 | 5.73 | 5.7 |
| # N's per 100kbp | 0 | 0 | 0 | 0 |
| Genome Statistics | ||||
| Genome fraction(%) | 99.945 | 99.756 | 99.98 | 99.775 |
| Duplication ratio | 1.001 | 1 | 1.002 | 1.001 |
| # genes | 4336 +2 part | 4316 +18 part | 4331 +8 part | 4320 +15 part |
| NGA50 | 2926354 | 1382073 | 1271789 | 1392434 |
| Running Time | 1hr 19m | 41m 3s | 32m 30s | 31m 16s |
without genomeSize and pbCNS (more detail)
| Statistics without reference | All Data | 6 SMRT cells : 1st Set | 6 SMRT cells : 2nd Set | 6 SMRT cells : 3rd Set |
| # contigs | 1 | 1 | 1 | 1 |
| Largest contig | 5164065 | 5163932 | 5163855 | 5163813 |
| Total length | 5164065 | 5163932 | 5163855 | 5163813 |
| N50 | 5164065 | 5163932 | 5163855 | 5163813 |
| Misassemblies | ||||
| # misassemblies | 0 | 1 | 0 | 0 |
| Misassembled contigs length | 0 | 5163932 | 0 | 0 |
| Mismatches | ||||
| # mismatches per 100kbp | 8.27 | 0.02 | 8.290 | 8.35 |
| # indels per 100kbp | 0.76 | 0.19 | 1.160 | 1.160 |
| # N's per 100kbp | 0 | 0 | 0 | 0 |
| Genome Statistics | ||||
| Genome fraction(%) | 99.922 | 99.942 | 99.918 | 99.917 |
| Duplication ratio | 1 | 1.001 | 1 | 1 |
| # genes | 4335 +3 part | 4336 +2 part | 4335 +3 part | 4335 +3 part |
| NGA50 | 5164065 | 2926240 | 5163855 | 5163813 |
| Running Time | 1hr 15m | 1hr 14m | 1hr 27m | 1hr 24m |
Dataset 9 (E. coli K-12, P4-C2 chemistry, 20 Kbp, 1 SMRT cell)
We used all SMRT cells and evaluated the assemblies by QUAST against the reference genome (NC_000913) and Ec_gene_list. (more detail)
PBcR -pbCNS -length 500 -partitions 200 -l ecoli -s pacbio.spec -fastq filtered_subreads.fastq PBcR -pbCNS -length 500 -partitions 200 -l ecoli -s pacbio.spec -fastq filtered_subreads.fastq genomeSize=4650000 PBcR -pbCNS -length 500 -partitions 200 -l ecoli -s pacbio.spec -fastq filtered_subreads.fastq genomeSize=4185000 PBcR -pbCNS -length 500 -partitions 200 -l ecoli -s pacbio.spec -fastq filtered_subreads.fastq genomeSize=5115000 PBcR -pbCNS -length 500 -partitions 200 -l ecoli -s pacbio.spec -fastq filtered_subreads.fastq genomeSize=3720000 PBcR -pbCNS -length 500 -partitions 200 -l ecoli -s pacbio.spec -fastq filtered_subreads.fastq genomeSize=5580000
Performance
with different genomeSize (more detail)
| Statistics without reference | genomeSize=4650000 | genomeSize=4185000 | genomeSize=5115000 | genomeSize=3720000 | genomeSize=5580000 |
| # contigs | 1 | 1 | 1 | 1 | 1 |
| Largest contig | 4644061 | 4651184 | 4644056 | 4651207 | 4651348 |
| Total length | 4644061 | 4651184 | 4644056 | 4651207 | 4651348 |
| N50 | 4644061 | 4651184 | 4644056 | 4651207 | 4651348 |
| Misassemblies | |||||
| # misassemblies | 8 | 8 | 8 | 8 | 8 |
| Misassembled contigs length | 4644061 | 4651184 | 4644056 | 4651207 | 4651348 |
| Mismatches | |||||
| # mismatches per 100kbp | 0.13 | 0.39 | 0.13 | 0.34 | 0.19 |
| # indels per 100kbp | 31.34 | 31.64 | 31.27 | 33.04 | 30.63 |
| # N's per 100kbp | 0 | 0 | 0 | 0 | 0 |
| Genome Statistics | |||||
| Genome fraction(%) | 100 | 99.998 | 100 | 99.998 | 100 |
| Duplication ratio | 1.001 | 1.003 | 1.001 | 1.003 | 1.003 |
| # genes | 4494 + 3 part | 4493 + 4 part | 4494 + 3 part | 4493 + 4 part | 4494 +3 part |
| NGA50 | 3025485 | 960375 | 3025483 | 960380 | 960403 |
| Running Time | 24m 50s | 23m 26s | 24m 25s | 21m 32s | 26m 5s |
without genomeSize (more detail)
| Statistics without reference | All Data |
| # contigs | 1 |
| Largest contig | 4651323 |
| Total length | 4651323 |
| N50 | 4651323 |
| Misassemblies | |
| # misassemblies | 8 |
| Misassembled contigs length | 4651323 |
| Mismatches | |
| # mismatches per 100kbp | 0.13 |
| # indels per 100kbp | 31.04 |
| # N's per 100kbp | 0 |
| Genome Statistics | |
| Genome fraction(%) | 100 |
| Duplication ratio | 1.003 |
| # genes | 4494+3 part |
| NGA50 | 960 398 |
| Running Time | 29m 13s |